Fig. 2.
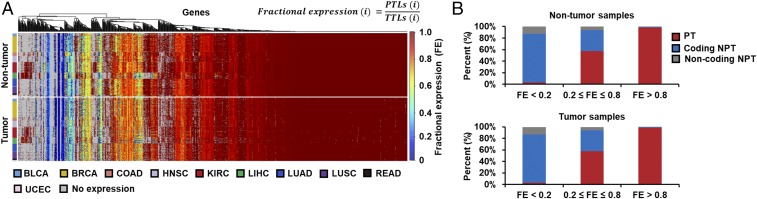
Relative expression levels of principal transcripts (PTs) in metabolic genes from 446 TCGA personal RNA-Seq data across the 10 cancer types. (A) Heat maps showing the distribution of FEs for all the metabolic genes from nontumor and tumor samples of the RNA-Seq data. FE is calculated by dividing principal transcript levels (PTLs) by total transcript levels (TTLs) for each metabolic gene, i. Metabolic genes in blue regions have high TTLs but low PTLs (i.e., low FE values), whereas metabolic genes in red regions have both consistent TTLs and PTLs (i.e., high FE values). Unexpressed genes are shown in gray. (B) Relative expression levels of PTs, coding NPTs, and noncoding NPTs for three different ranges of FEs in the nontumor and tumor samples.