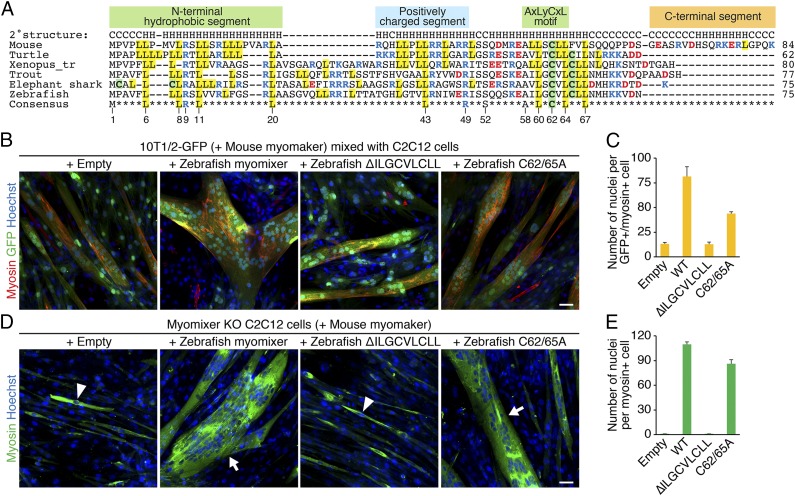
Fig. 4.
Functional analyses of zebrafish myomixer mutant proteins. (A) Cross-species alignment of myomixer proteins. Basic residues are shown in blue, and acidic residues are shown in red. Cysteines are highlighted in green, and leucines are highlighted in yellow. Identical residues in all orthologs are shown at the bottom. The numbers below the consensus sequence refer to the amino acid positions in zebrafish myomixer. (B) The 10T1/2-GFP fibroblasts were infected by retroviruses expressing mouse myomaker with or without wild-type or mutant zebrafish myomixer and mixed with C2C12 cells for heterologous fusion following 1 wk of differentiation. Cells were stained with anti-myosin and Hoechst. (C) Quantification of the fusion index in B. (D) Myomixer KO C2C12 cells expressing mouse myomaker with or without wild-type or mutant zebrafish myomixer were induced to fuse in DM for 1 wk. Cells were stained with anti-myosin and Hoechst. (E) Quantification of the fusion index in D. Arrowheads indicate mononucleated cells; arrows indicate multinucleated cells. (Scale bars, 50 µm.)