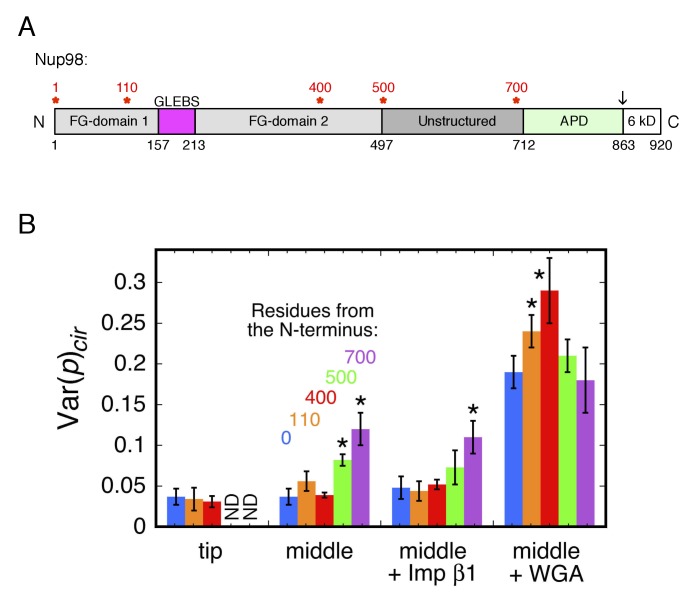
Figure 5. Rotational mobility of mEos3 at different positions within the Nup98 disordered domains.
(A) Human Nup98 domain structure. The GLEBS domain is a binding motif for RNA export factors. The autoproteolytic domain (APD) co-translationally cleaves the C-terminal 6 kDa domain after residue 863 (arrow) (Rosenblum and Blobel, 1999). Amino acids numbers are indicated on the bottom and stars (*) indicate the positions at which the mEos3 probe was incorporated (0, 110, 400, 500, 700 residues from the N-terminus). The regions from residues 1–157 and 213–712 are considered disordered. Adapted from (Ren et al., 2010). (B) Var(p)cir values obtained under the indicated conditions for the mEos3 probe at different positions within the Nup98 disordered domains (see A). p-PALM data were obtained as in Figure 3. The stars (*) indicate significantly different values from the wild-type (blue) condition within the same group according to a two-sided Welch’s t-test (95%). Note that the actual significance level is expected to be higher than indicated since the error bars are wider than a typical standard deviation (see Figure 2—figure supplement 6).