Extended Data Figure 3. Epigenetic and transcriptional regulation of normal CD8 differentiation.
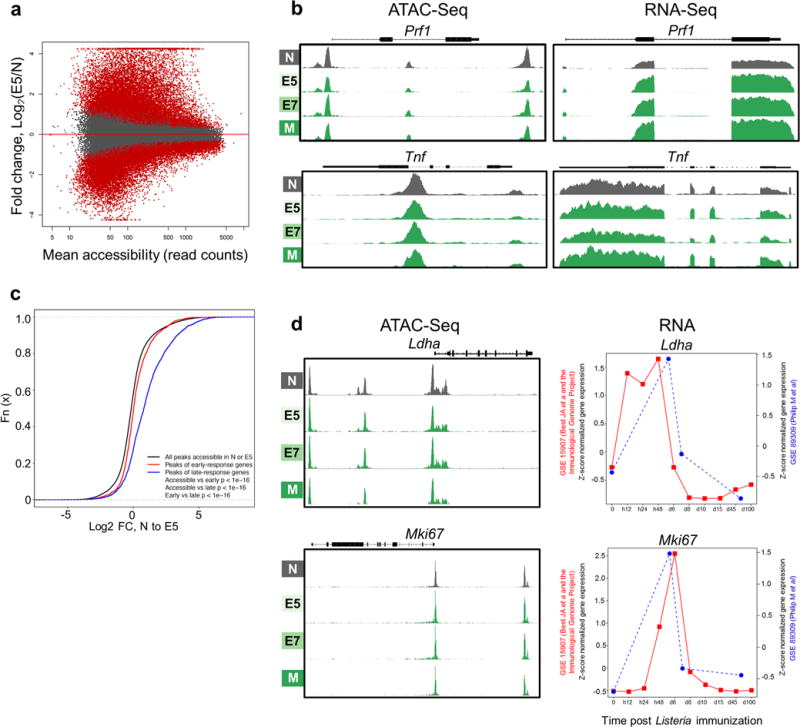
a, ATAC-Seq data reveals massive chromatin remodeling during normal CD8 T cell differentiation. MA plot of Naïve (N) and Day 5 Effectors (E5) showing log2 ratios of peak accessibility (E5/N) versus mean read counts for all atlas peaks. Significantly differentially accessible peaks are shown in red (FDR < 0.05). b, Epigenetic and transcriptional regulation of CD8 effector genes. ATAC-seq (left) and RNA-seq (right) signal profiles of Prf1 and Tnf in Naïve (N), Effectors (E5 and E7), and Memory (M) TCRTAG during acute LmTAG infection. (c – d), Epigenetic and transcriptional regulation of early CD8 response genes in TCRTAG during acute listeria infection. Published expression data from the Immunological Genome Project (JA Best et al, Nat Imm (2013); GSE 15907) were used; early-response genes increase in expression within the first 12–24 hours and late-response genes increase expression 24–48 hours after naïve T cells encounter LmOVA as determined by Best JA et al. c, Cumulative distribution function of peak accessibility changes between N and E5. Peaks associated with early-response genes show fewer changes in accessibility as compared to peaks associated with late-response genes. The black line shows all peaks accessible in N or E5, the red line shows peaks associated with early-response genes and the blue line shows peaks associated with late-response genes. d, ATAC-seq signal profiles (left) and RNA expression (right) of the early response genes Ldha (top) and Mki67 (bottom) in N, E5/E7, and M TCRTAG during acute LmTAG infection (blue line; GSE 89309; current dataset Philip M et al.) overlaid with expression data from Best JA et al./Immunological Genome Project (red line).
