Figure 2. RAS induces RPL23 expression.
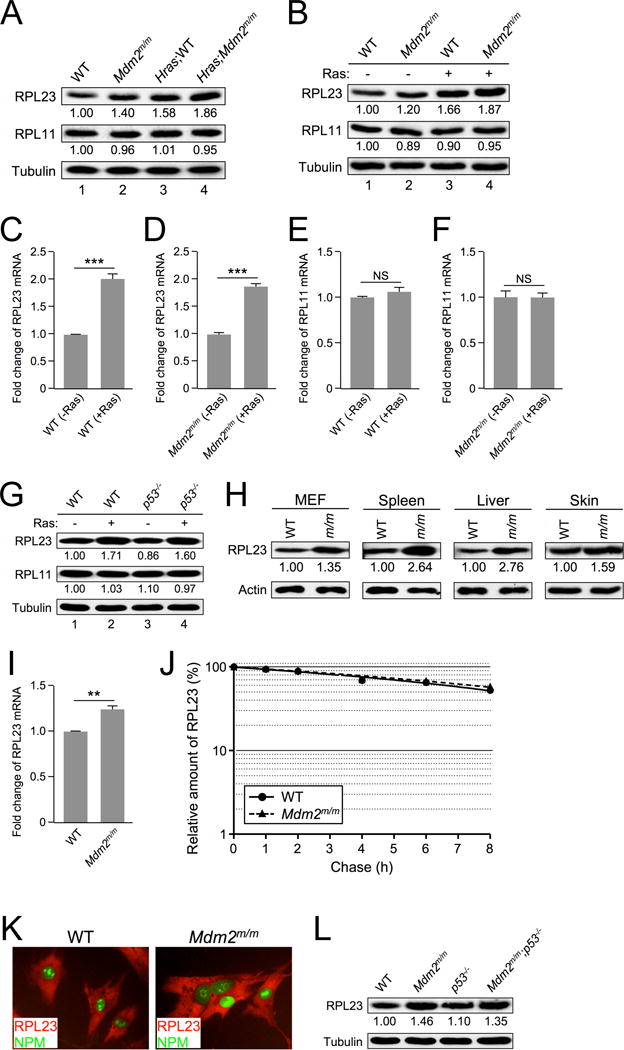
A. Extracts from skin tissue of non-tumor-bearing WT and Mdm2m/m mice and from their Hras transgenic counterparts were analyzed by western blot. The relative expression of RPL23 and RPL11 is shown under the blot (analyzed by ImageJ software, 1.47v).
B. Early passage WT and Mdm2m/m MEFs were infected with retrovirus expressing either pBabe vector (−) or pBabe-HRASG12V (+), selected in puromycin for three days, then allowed to recover for 48 hours before harvesting for western blot analysis. The relative expression of RPL23 and RPL11 is shown under the blot (analyzed by ImageJ software, 1.47v).
C. WT MEFs were infected with retrovirus expressing either pBabe vector (-Ras) or pBabe-HRASG12V (+Ras) and harvested for qRT-PCR mRNA analysis. Relative RPL23 mRNA expression was calculated using β-GAPDH as an internal control. Data are represented as mean ± SEM, and were analyzed by Student’s t test. (*** indicates p value <0.001)
D. Mdm2m/m MEFs were infected with retrovirus expressing either pBabe vector (−Ras) or pBabe-HRASG12V (+Ras) and harvested for qRT-PCR mRNA analysis. Relative RPL23 mRNA expression was calculated using β-GAPDH as an internal control. (*** indicates p value <0.001)
E. WT MEFs were infected with retrovirus expressing either pBabe vector (−Ras) or pBabe-HRASG12V (+Ras) and harvested for qRT-PCR mRNA analysis. Relative RPL11 mRNA expression was calculated using β-GAPDH as an internal control. (NS indicates no statistically significant difference between samples)
F. Mdm2m/m MEFs were infected with retrovirus expressing either pBabe vector (−Ras) or pBabe-HRASG12V (+Ras) and harvested for qRT-PCR mRNA analysis. Relative RPL11 mRNA expression was calculated using β-GAPDH as an internal control. (NS indicates no statistically significant difference between samples)
G. Early passage WT and p53−/− MEFs were infected with retrovirus expressing either pBabe vector (−) or pBabe-HRASG12V (+), selected in puromycin for three days, then allowed to recover for 48 hours before harvesting for western blot analysis. The relative expression of RPL23 and RPL11 is shown under the blot (analyzed by ImageJ software, 1.47v).
H. Extracts from WT and Mdm2m/m MEFs and from tissues of 30-week-old WT and Mdm2m/m mice were analyzed by western blot. The relative expression of RPL23 is shown under the blot (analyzed by ImageJ software, 1.47v).
I. Early passage WT and Mdm2m/m MEFs were harvested for qRT-PCR mRNA analysis. Relative RPL23 mRNA expression was calculated using β-GAPDH as an internal control.
J. Half-life assay of RPL23 was carried out using early passage (P1) WT and Mdm2m/m MEFs treated with cycloheximide (50 μg/mL) and harvested with SDS lysis buffer at the indicated time points. The amount of RPL23 was quantified by densitometry, normalized to the level of actin, and plotted.
K. Early passage WT and Mdm2m/m MEFs were fixed and stained with rabbit anti-RPL23 antibody and a fluorescein isothiocyanate-conjugated anti-rabbit secondary antibody (red color), and mouse anti-B23 (NPM) antibody and a fluorescein isothiocyanate-conjugated anti-mouse secondary antibody (green color). Fluorescence images were captured with a cooled charge-coupled device color digital camera (Model 2.2.0, Diagnostic) on an Olympus IX81 inverted microscope equipped with the appropriate fluorescence filters.
L. Extracts from WT, Mdm2m/m, p53−/−, and Mdm2m/m;p53−/− MEFs were analyzed by western blot. The relative expression of RPL23 is shown below the blot (analyzed by ImageJ software, 1.47v).
