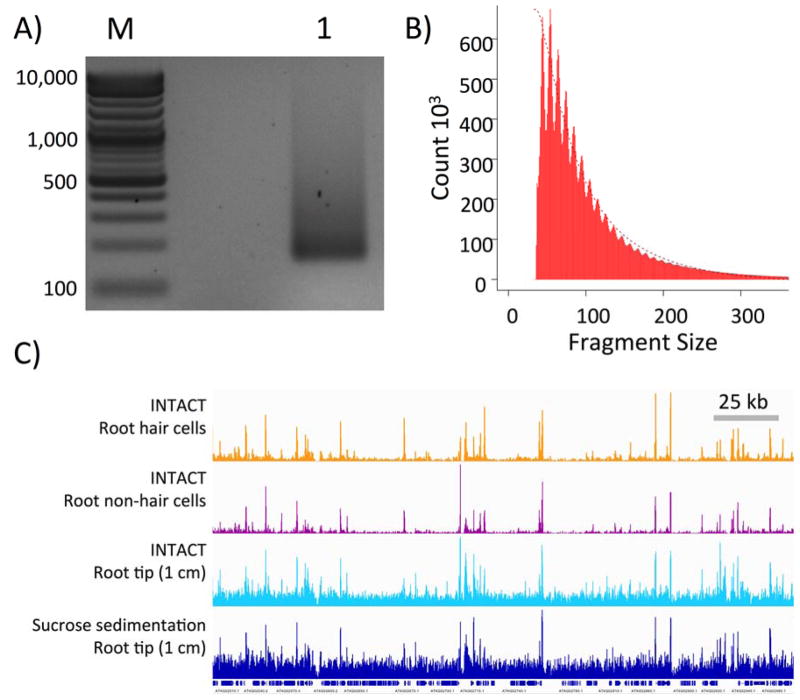
Figure 2. ATAC-seq library preparation and high-throughput sequencing.
A) An amplified ATAC-seq library purified with Ampure XP beads (lane “1”) was resolved in a 2% agarose gel stained with ethidium bromide. Lane “M” is the molecular weight marker lane. Amplified library fragments generally range in size from 180 bp to several kb in size. The size distribution of the resolved gel may vary somewhat, but the final product should be free of adapter dimers (distinct band around 125 bp) and primer dimers (distinct band around 80 bp). See Note 11. B) Insert sizes of ATAC-seq paired-end reads from 50,000 nuclei isolated by INTACT from non-hair cells calculated using the InsertSizeMetrics option from Picard Tools (Note 13). The distribution shows periodicity of helical pitch of DNA for fragments smaller than 200 bp. Fragments containing one or more nucleosomes, related to insert periodicity increasing in 150 bp, were not observed using the transposase:nuclei and bead:DNA ratios described in this protocol. C) Integrated Genome Viewer snapshot of four different libraries sequenced on the Illumina platform. The tracks shown are of ATAC sequencing reads from INTACT isolated nuclei from root hair cells (orange), root non-hair cells (purple), root tip (cyan), and sucrose sedimentation isolated nuclei from 1 cm root tip (navy). Gene tracks are shown below the ATAC-seq tracks and a 25 kb scale bar is show