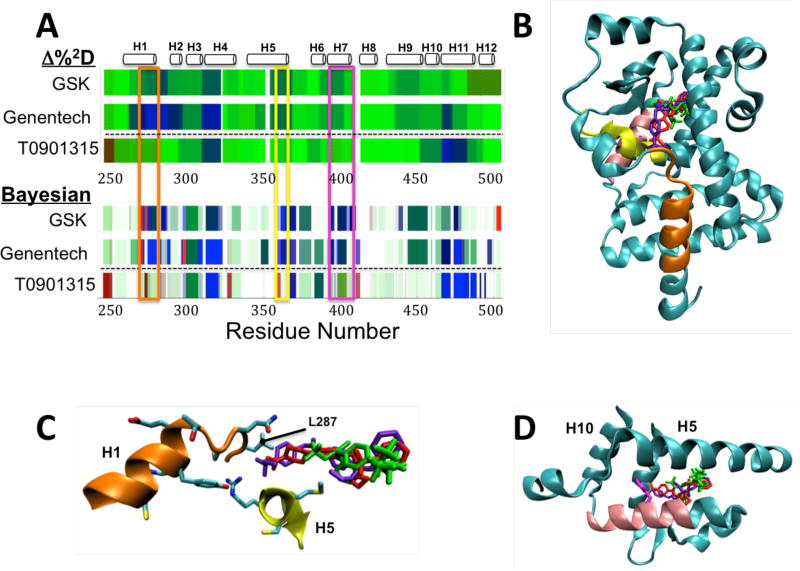
Figure 4. ΔHDX of ligands of RORγ.
A) ΔHDX of agonists GSK and Genentech and inverse agonist T0901315 with respect to RORg as analyzed by the Δ%2D method (top) and Bayesian method (bottom). Areas of major difference between the two classes of compound are seen in residues 276 to 287 (orange box), 365 to 373 (yellow box) and residues 399 to 410 (pink box). B) Visualization of the three ligands bound to RORg, T0901315 (green, PDB: 4NB6), GSK (red, PDB: 4NIE) and Genentech (blue, PDB: 4WPF) on the structure of RORγ (from PDB 4NIE). Areas of peptide with major ΔHDX differences highlighted in part A are colored. C) Close up of orange and yellow areas and the side chain interactions with the ligand binding site, showing the difference in interactions in this area between T0901315 (green) and the two agonists. D) Close up of pink region in helix 7 with the side chain of F403 in magenta showing its positioning just outside the ligand binding site at the junction of helices 5 and 10.