Figure 7. Functional binding of unpaired DNA and RNA to DDX3X and Ded1p.
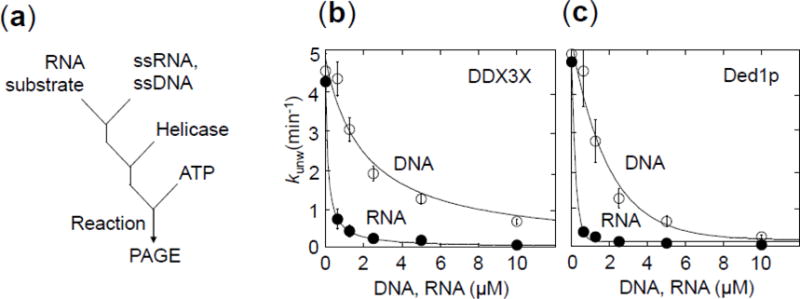
(a) Reaction scheme of the competition reaction to measure functional binding of unpaired DNA and RNA to DDX3X and Ded1p.
(b) Unwinding reactions with DDX3X (150 nM, ATP: 2 mM) RNA substrate: 16 bp duplex RNA substrate with 25 nt 3′ tail, 0.5 nM) with increasing concentrations of a 41 nt DNA (open circles) and a 41 nt RNA (closed circles). Data points represent averages of at least three independent measurements, error bars mark one standard deviation. Functional affinities Ki for unpaired DNA and RNA were calculated as in panel (b) (KiDNA = 3,560 ± 100 nM, KiRNA = 71 ± 5 nM).
(c) Unwinding reactions with Ded1p (150 nM, ATP: 2 mM). RNA substrate: 16 bp duplex RNA substrate with 25 nt 3′ tail, 0.5 nM) with increasing concentrations of a 41 nt DNA (open circles) and a 41 nt RNA (closed circles). Data points represent averages of at least three independent measurements, error bars mark one standard deviation. The lines mark a trend. Functional affinities could not be calculated because the unwinding rate constant at enzyme saturation without inhibitor could not be determined (Fig.3b).
