Figure 9. Unwinding with different nucleoside triphosphates.
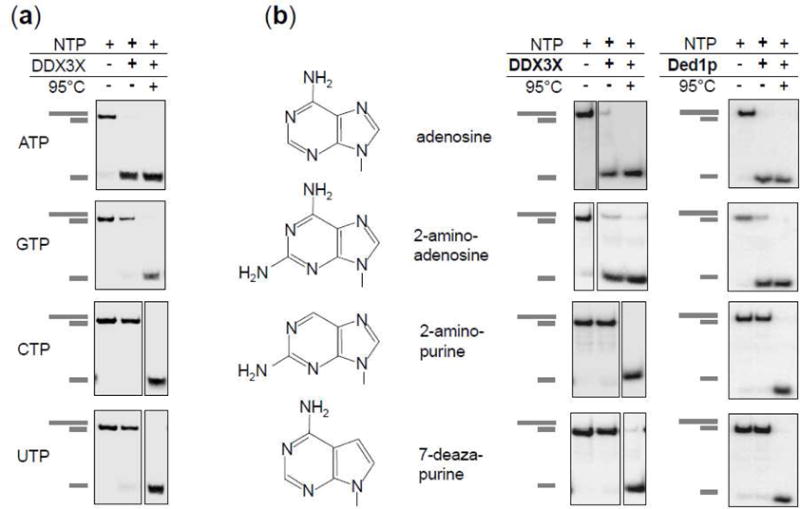
(a) Representative unwinding reactions by DDX3X (150nM) with ATP, GTP, CTP and UTP (2 mM each). (RNA: 16 bp, 25 nt 3′ tail, 0.5 nM reaction time: 15 min). Cartoons on the left mark duplex RNA substrate and unwound product.
(b) Unwinding with ATP, 2-amino adenosine triphosphate, 2-amino purine triphosphate, and 7-deaza purine triphosphate. Chemical structures of the nucleobases are depicted. Gel panels show representative unwinding reactions by DDX3X (left column) (150 nM) and Ded1p (right column, 150nM). (RNA: 16 bp, 25 nt 3′ tail, 0.5 nM, reaction time: 15 min). Cartoons on the left mark duplex RNA substrate and unwound product.
