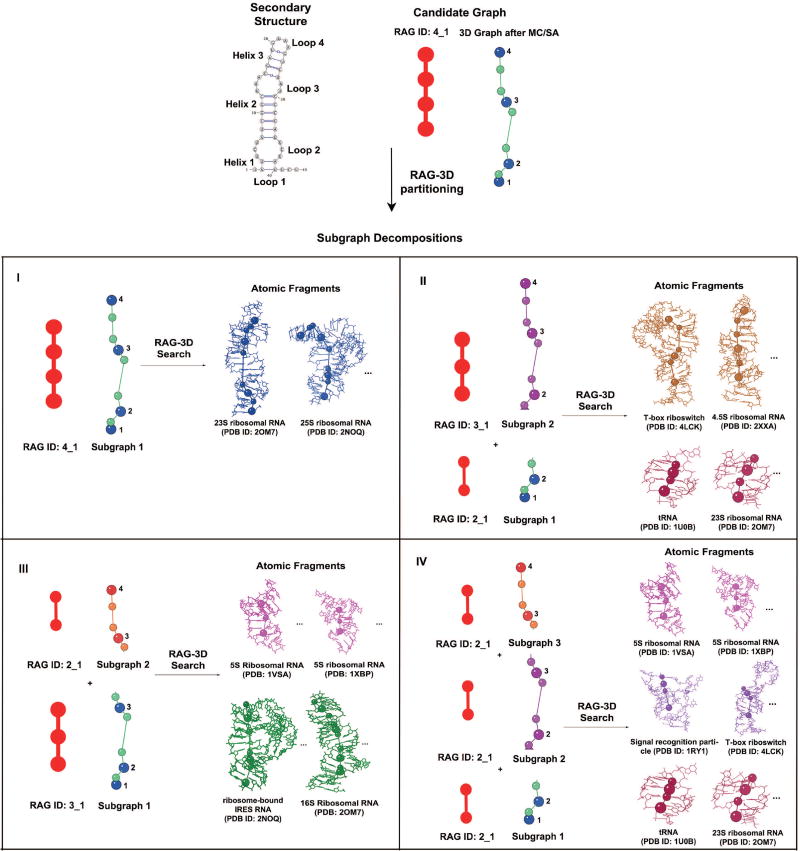
Figure 7. Sample F-RAG Input for the pentanucleotide AUUCU repeat expansion RNA (PDB ID: 5BTM).
The 2D structure, candidate graph, corresponding subgraphs, and associated atomic fragments from the RAG-3D search that serve as input to F-RAG are shown. For this 4 1 target, we obtain 4 subgraph decompositions as shown. For each subgraph decomposition, we run F-RAG using the 10 lowest graph RMSD atomic fragments for each target subgraph, to obtain many atomic models. We then select all atomic models that have the same number of residues as the target structure (or the highest number of residues in case of missing residues), sort them in increasing order of their score (based on our knowledge-based statistical potential), and select the top scoring models for geometry optimization with PHENIX. In Figure 8, we illustrate the steps of F-RAG for one subgraph decomposition, namely III.