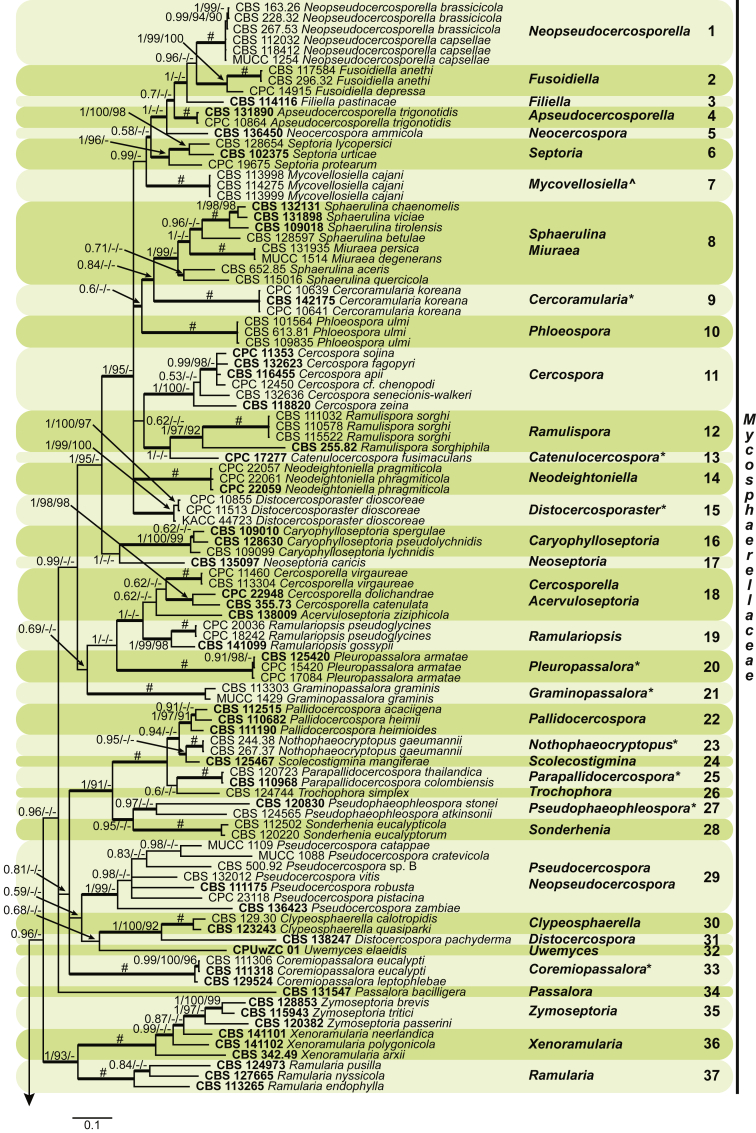
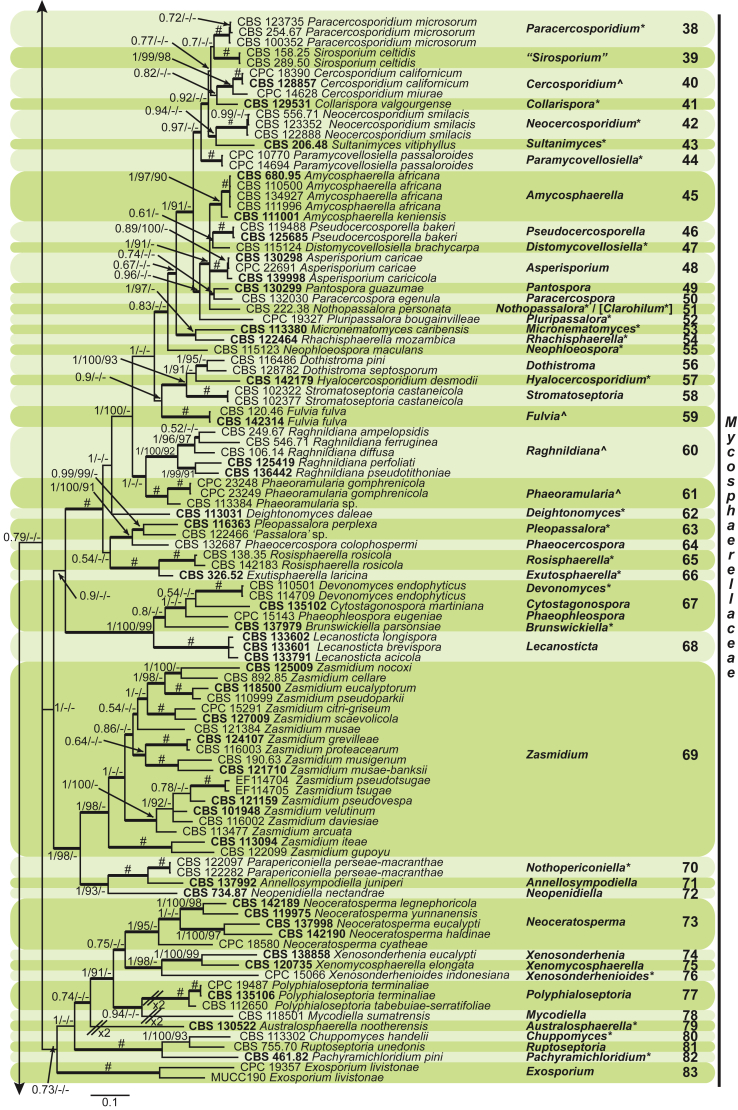
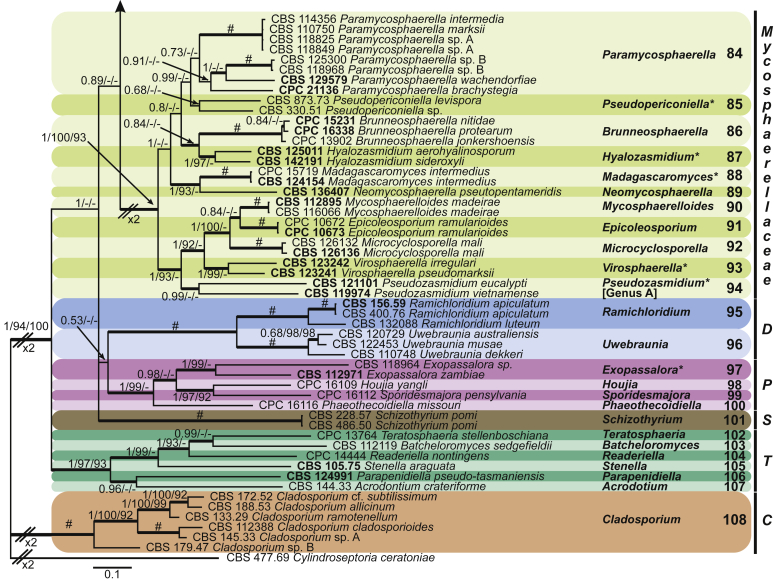
Fig. 1.
Phylogenetic tree (50 % majority rule consensus) resulting from a Bayesian analysis of the combined LSU and rpb2 sequence alignment (dataset 1). Bayesian posterior probabilities (PP), maximum likelihood bootstrap support values (≥ 90 %; ML-BS) and maximum parsimony bootstrap support values (≥ 90 %; MP-BS) are indicated at the nodes (PP/ML-BS/MP-BS; a hash (#) symbol denotes fully-supported branches) and the scale bar represents the expected number of changes per site. Branches in a thicker stroke represent the branches present in the strict consensus parsimony tree. Genera clades are delimited in coloured boxes, with the genus name and clade number indicated to the right. All taxa names are written in black, ex-type strains are represented in bold, novel genera denoted with an asterisk (*) and resurrected genera with a circumflex (ˆ). A vertical bar is used to the right of the coloured boxes and encompasses all genera within their respective families. The family name Mycosphaerellaceae is unabbreviated while the rest are abbreviated as follows: D = Dissoconiaceae, P = Phaeothecoidiellaceae, S = Schizothyriaceae, T = Teratosphaeriaceae, C = Cladosporiaceae. The tree was rooted to Cylindroseptoria ceratoniae (CBS 477.69).