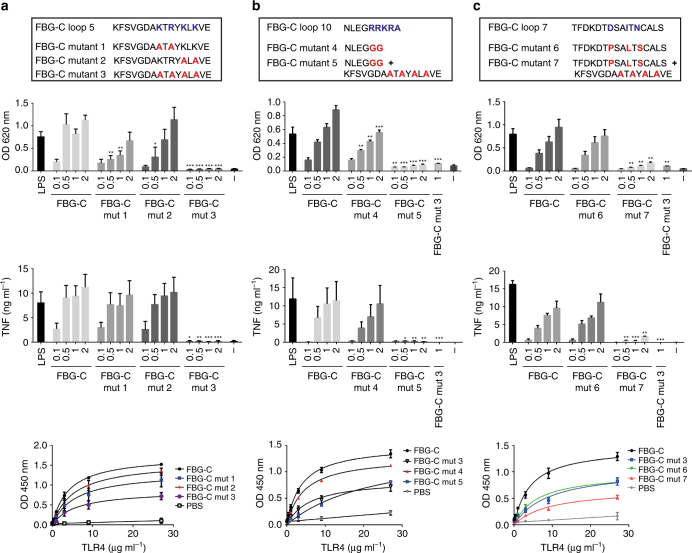
Fig. 4.
Pinpointing amino acids in loops 5, 7, and 10 of FBG-C that mediate TLR4 binding and activation. Upper panel: Sequences of wild-type FBG-C and mutants 1–7, highlighting wild-type amino acids in blue and mutations in red (loop 5 variants are shown in a, loop 10 in b, and loop 7 in c). Second panel: THP1 NF-kB cells were left unstimulated (−) or stimulated for 24 h with LPS (1 ng ml−1), increasing doses (μM) of FBG-C or FBG-C mutants 1–7. NF-kB activation was measured using QUANTI-Blue™. Data shown as mean ± SEM. n = 4 independent experiments. Paired t-test vs. FBG-C, *p < 0.05, **p < 0.01, ***p < 0.001. Third panel: Primary human macrophages were left unstimulated (−) or stimulated for 24 h with LPS (1 ng ml−1), increasing doses (μM) of FBG-C or FBG-C mutants 1–7. Cytokines synthesis was measured by ELISA. Data shown as mean ± SEM. n = 4 independent donors. Paired t-test vs. FBG-C, *p < 0.05, **p < 0.01, ***p < 0.001. Bottom panel: 96-well plates were coated with 1 µg ml−1 of FBG-C or FBG-C mutants 1–7, and TLR4 was added in a dose-dependent manner. Curves were fitted in GraphPad Prism using one-binding site hyperbola equation. Data shown as mean ± SEM; n = 4