Figure 3.
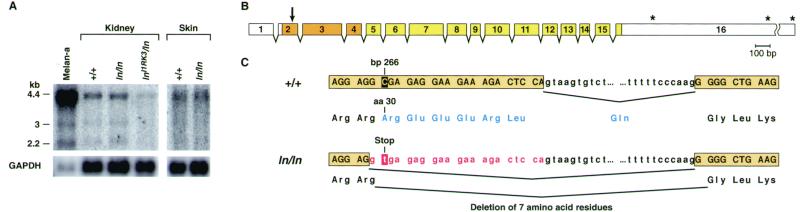
The ln mutation results from a defect in Mlph. (A) Northern blot analysis of Mlph expression in wild-type (C57BR/cdJ), C57L/J-ln/ln, and In(1)1Rk-lnllRK3/ln tissues. Transcript sizes (in kb) are depicted on the left. The amount and size of Mlph transcripts seem unchanged in ln/ln mutants, but are reduced in lnllRK3/ln tissue. Also shown is Mlph expression in wild-type melan-a melanocytes (27). Note that Mlph is expressed at high levels in melan-a cells. (B) The intron/exon organization of the mouse Mlph gene. Coding regions are shown in color and untranslated region (UTR) sequences in white. The asterisks indicate the locations of three alternatively used polyadenylation sites, orange boxes represent the conserved Rab-effector domain, and the arrow denotes the site of the ln mutation. (C) A C-to-T transition at bp 266 (shaded) results in altered splicing between exons 2 and 3. The ln mutation generates a new splice donor site, so that the last 21 bp (shown in red) of exon 2 are spliced out of the mature message. If this message were translated, it would produce a protein that lacks amino acid residues 30–36 (REEERLQ), shown in blue. If there were some readthrough of the mutant message, a stop codon at amino acid 30 would severely truncate the protein.