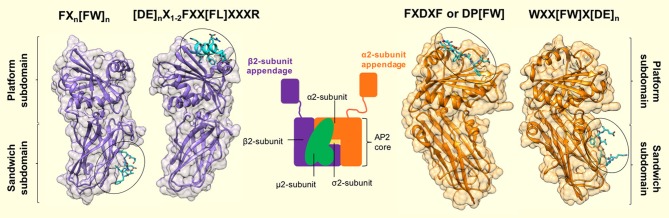
Figure 4.
Motif binding to the appendage domains of AP2. The α2- and β2-subunit appendages of AP2 (orange and purple, respectively) share a similar bilobal structure each consisting of an N-terminal sandwich subdomain attached to a C-terminal platform subdomain. Each subdomain, of each appendage contains a distinct interaction surface for protein partner binding, which results in a single AP2 molecule possessing 4 separate contact sites. Specificity for each site is conferred by short, interaction motifs which are characterized by aromatic side chains. The sandwich subdomain of the α2-appendage binds to WXX[FW]X[DE]n-containing ligands (where X is any amino acid). [PDB code: 1W80, (Praefcke et al., 2004)]. The platform subdomain of the same appendage binds either FXDXF or DP[FW] motifs [PDB code: 1KY7, (Brett et al., 2002)]. The same subdomain of the β2-subunit binds to [DE]nX1−2FXX[FL]XXXR sequences that are presented in a α-helical conformation. Such motifs are present in β-arrestin, ARH and epsins (Edeling et al., 2006; Schmid et al., 2006) [PDB code: 2G30, (Edeling et al., 2006)]. Finally – the sandwich subdomain binds to a Phe-rich motif that is present in proteins, Eps15 and AP180 [PDB code: 2IV9, (Schmid et al., 2006)]. Peptide motif nomenclature as in Traub (2009).