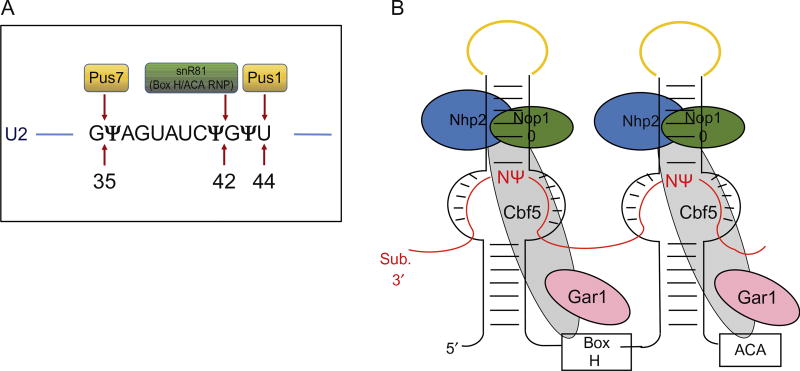
Fig. 3.
Schematic of uridine-to-pseudouridine conversion catalyzed by two mechanisms in Saccharomyces cerevisiae. (A). The pseudouridylation catalyzed by a standalone protein pseudouridine synthase 1 (Pus1p) and pseudouridine synthase 7 (Pus7p) in conversion of U44 to Ψ44 and U35 to Ψ35, respectively, in U2 snRNA. Pseudouridylation at position 42 by snR81 box H/ACA RNP is also indicated. (For the structure of a pseudouridine synthase, refer to chapter “The evolution of multi-substrate specificity by RNA modification enzymes” by Alfonzo) (B). The box H/ACA RNP, consisting of a small noncoding box H/ACA guide RNA that folds into a typical hairpin–hinge–hairpin–tail secondary structure forming two hairpins and four common core proteins. The core proteins are centromere-binding factor 5 (Cbf5; known as dyskerin/NAP57 in mammals), glycine–arginine-rich protein 1 (Gar1), nonhistone protein 2 (Nhp2), and nucleolar protein 10 (Nop10). The substrate RNA (indicated in red lines) drapes around the box H/ACA RNP complex via complementary base-pairing interactions with the guiding pockets of box H/ACA RNA, positioning the target uridine, and its 3′ adjacent nucleotide (N) at the base of the upper stem and leaving them unpaired. The apical loops of box H/ACA RNA that hold the CAB boxes are indicated in yellow.