Figure 3. Identification of a conserved SE sub-domain essential for Ly6Clow Mo development.
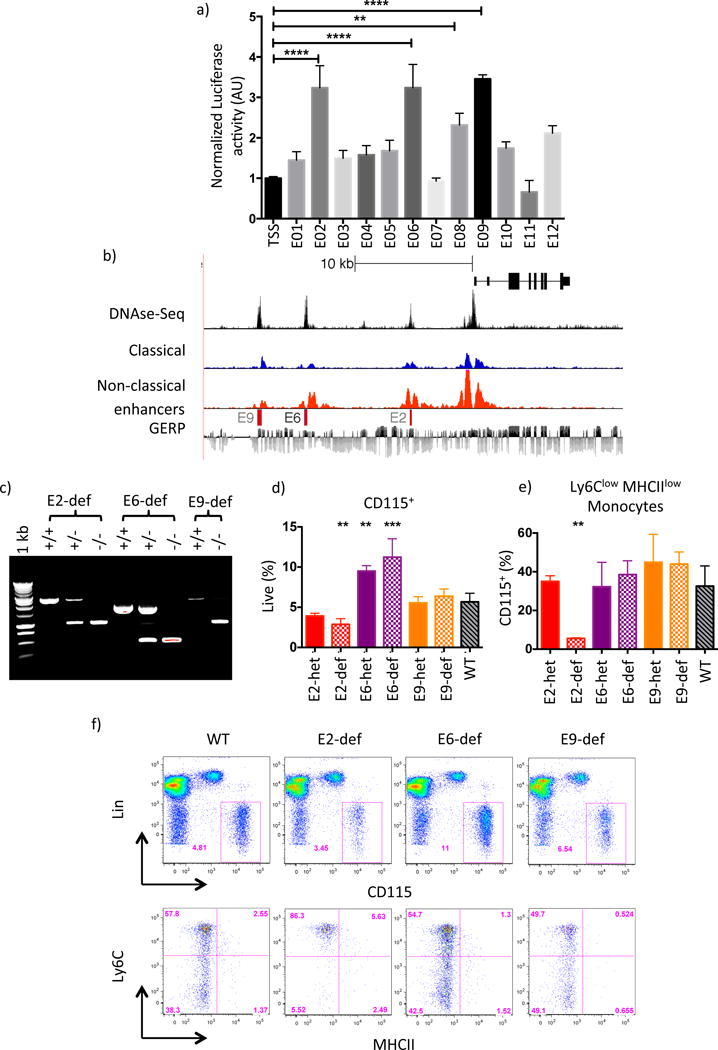
a) Relative luciferase activity in RAW264.7 cells of candidate enhancer regions cloned into pGL4.Nr4a1 vector. Results shown are averaged over 2 independent experiments. Error bars represent SD, P-values calculated by 2-way anova * P<0.05, **P<0.01, ****P<0.0001. b) UCSC genome browser screenshot of the human Nr4a1 locus. H3K27ac tracks for CD14+CD16dim (classical) and CD14dimCD16+ (non-classical) Mo are shown alongside BLAT alignments of nucleosome-free DNA sequence obtained from mouse E2, E6 and E9. Mo DNase-Seq data were taken from (Boyle et al, 2008); nucleotide sequence conservation is indicated using the GERP track. c) PCR Genotyping of E2, E6 and E9 heterozygous (het) and deficient (def) mice. d) and e) Enumeration of peripheral blood Mo in WT, E2, E6 and E9 domain-heterozygous (het) and -deficient (def) mice. Parametric t-tests performed relative to WT, ** p< 0.01 *** p<0.001. f) FACS gating and representative flow plot for each genotype in d) and e). Please refer to supplementary figure S4.
