FIGURE 2.
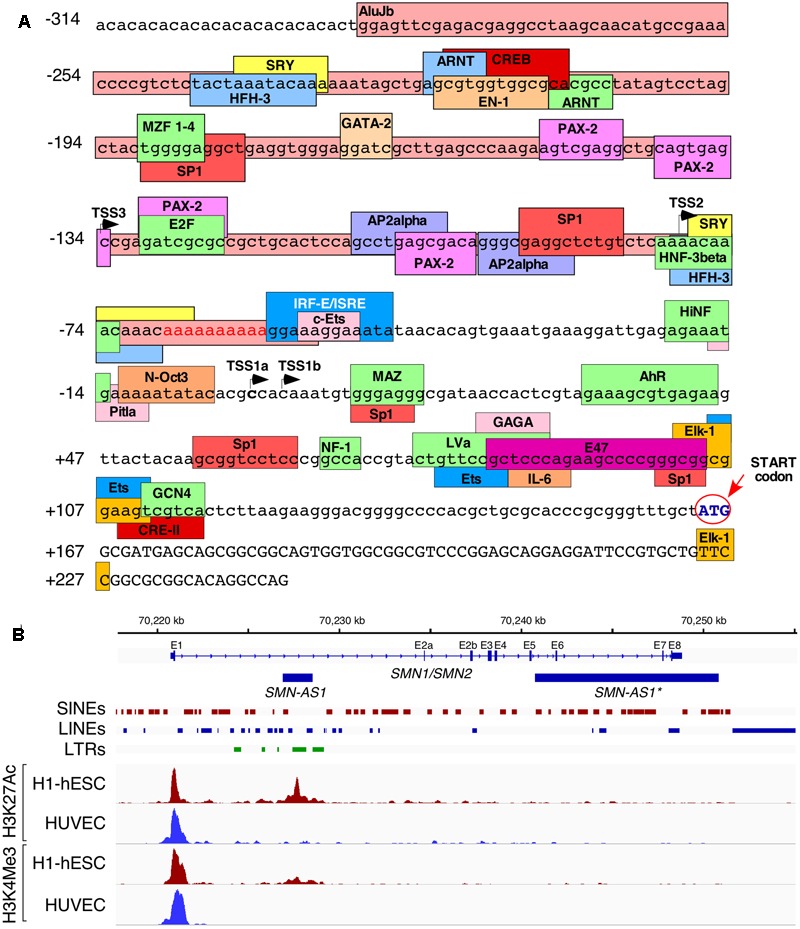
Alu repeats reshape transcriptional regulation of SMN. (A) Overview of the promoter region of SMN. Lowercase letters indicate untranscribed or untranslated regions, uppercase letters indicate coding sequences. A partial AluJb element is inserted from 289 to 54 bases upstream of the canonical TSS (TSS1a) and is indicated with a dark pink colored box. Other colored boxes indicate the locations of putative promoter elements and/or transcription factor binding sites. TSSs are indicated with black arrows. The start codon is written in blue and is indicated with a red arrow and circle. Promoter elements were either described previously (Singh et al., 2016) or were computationally predicted by ConSite (consite.genereg.net). (B) Locations of Polycomb-associated antisense transcripts derived from SMN. A genomic view of the SMN gene is shown. Location of SMN exons (E1–E8) and antisense transcripts are indicated with blue boxes, introns are indicated by lines with arrows indicating the direction of transcription. The locations of SINEs, LINEs, and LTRs are indicated. Four ChIP-Seq outputs are given below. The red peaks indicate the read depth from ChIP-Seq of human embryonic stem cells using antibodies against acetylated H3K27 (upper) or tri-methylated H3K4 (lower). The blue peaks indicate the same readout from umbilical cord-derived HUVEC cells. ChIP-Seq data was obtained from ENCODE and previously described by d’Ydewalle et al. (2017).
