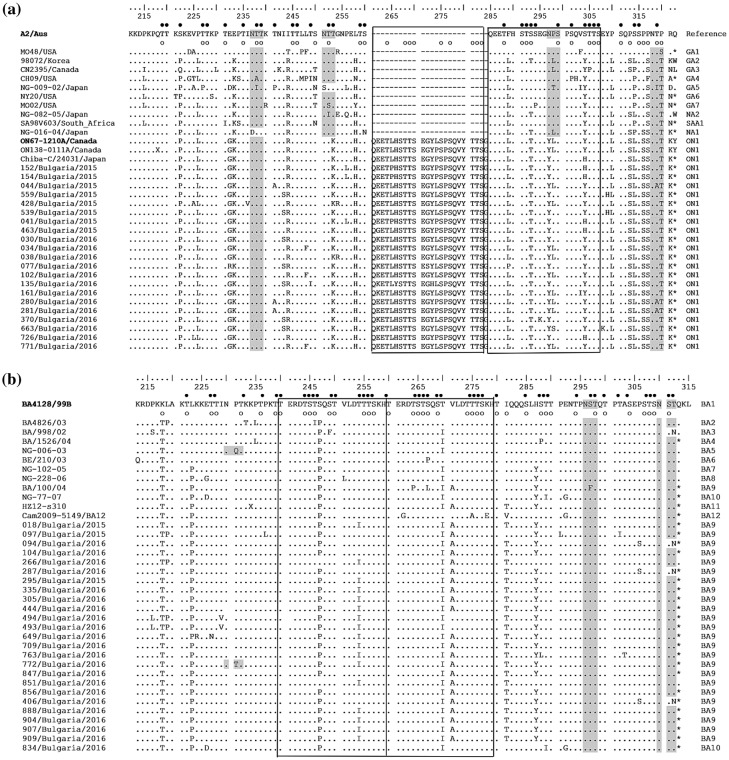
Figure 4.
Deduced amino acid alignments of the second variable region of the G-protein from RSV-A (a) and RSV-B (b) strains. The alignment (a) is shown relative to the sequence of reference RSV-A strain A2 (GenBank accession number M11486) and the alignment (b) is shown relative to the sequence of prototype BA strain BA4128/99B (GenBank accession number AY333364). The amino acid numbers correspond to G protein positions 212 to 298 of the strain A2 (a) and to G protein positions 213 to 315 of the strain BA4128/99B (b). Identical residues are identified as dots. Dashes indicate gaps corresponding to the nucleotide insertions; asterisks indicate the stop codons. The outlined rectangles represent the two copies of the duplicated 23-amino acid region in RSV-A strains (a) and the two copies of the duplicated 20-amino acid region in RSV-B strains (b). Genotype names are shown on the right. Light grey shading highlights the predicted N-glycosylation sites. Black circles denote the predicted O-glycosylation sites of the A2 reference strain (a) and BA prototype strain BA4128/99B (b); predicted O-glycosylation sites of Bulgarian strains are indicated by unfilled circles.