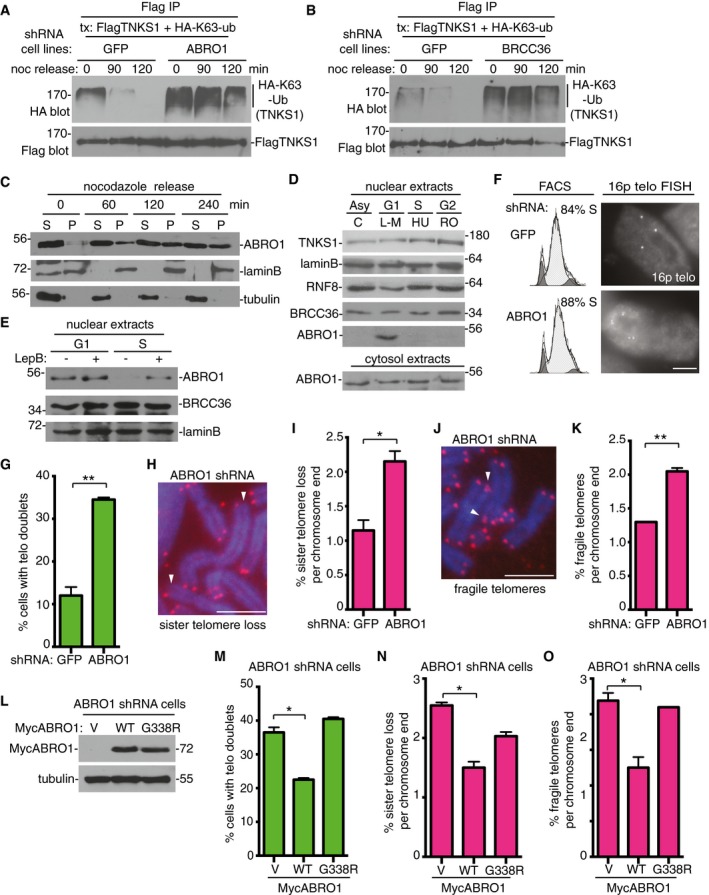
Figure 6. ABRO1 is required to remove K63‐Ub chains from tankyrase 1 in G1 to prevent premature resolution of sister telomere cohesion.
-
AABRO1 is required for removal of K63‐Ub chains from tankyrase 1 as cells exit mitosis and enter G1. GFP or ABRO1 shRNA HTC75 cell lines were transfected with FlagTNKS1 and HA‐K63‐Ub and treated with nocodazole for 20 h. Cells were isolated by mitotic shake‐off, replated, and harvested at 0, 90, and 120 min. Cell extracts were immunoprecipitated with anti‐Flag antibody and analyzed by immunoblotting with anti‐HA or Flag antibodies.
-
BBRCC36 is required for removal of K63‐Ub chains from tankyrase 1 as cells exit mitosis and enter G1. GFP or BRCC36 shRNA HTC75 cell lines were processed as described in (A).
-
CABRO1 enters the nuclear fraction as cells exit mitosis and enter G1. Following 20‐h nocodazole treatment, HTC75 cells were isolated by mitotic shake‐off, replated, harvested at 0, 60, 120, and 240 min, separated into nuclear pellet (P) and cytosolic supernatant (S) fractions, and analyzed by immunoblotting with the indicated antibodies.
-
DABRO1 localizes to the nuclear fraction in G1 phase of the cell cycle. HTC75 cells were synchronized by addition of L‐mimosine (G1), hydroxyurea (S), or R03306 (G2) for 18–20 h, harvested, separated into nuclear and cytosolic fractions, and analyzed by immunoblotting with the indicated antibodies.
-
EABRO1 is exported from the nucleus in S phase. HTC75 cells were synchronized in G1 or S (as above) and leptomycin B was added 3 h prior to harvest. The nuclear fractions were analyzed by immunoblotting with the indicated antibodies.
-
F, GTelomere cohesion is resolved prematurely in ABRO1‐depleted cells. (F) GFP or ABRO1 HTC75 shRNA cell lines were synchronized by a double thymidine block and analyzed by FACS and telomere FISH with a telomere 16ptelo probe 4 h after release from the second thymidine block. Scale bar, 5 μm. (G) Graphical representation of the frequency of cells with telomere doublets in S phase. Average of two independent experiments (n = 60 cells each) ± SEM. **P ≤ 0.01; Student's unpaired t‐test.
-
H–KABRO1 depletion leads to (H and I) sister telomere loss and (J and K) fragile telomeres. (H and J) Telomere FISH analysis with a (CCCTAA)3 repeat probe (red) of metaphase spreads from GFP or ABRO1 shRNA HTC75 cell lines. DNA was stained with DAPI (blue). Scale bars, 5 μm. Arrows indicate (H) sister telomere loss or (J) fragile telomeres. (I and K) Graphical representation of the frequency of (I) sister telomere loss or (K) or fragile telomeres per chromosome end. Average of two independent experiments (n = 798–846 chromosome ends each) ± SEM. *P ≤ 0.05, **P ≤ 0.01; Student's unpaired t‐test.
-
L–OThe telomere dysfunction in ABRO1‐depleted cells is rescued by ABRO1 WT, but not by the ABRO1 G338R tankyrase‐binding mutant. ABRO1 HTC75 shRNA cells were transfected with vector (V) or MycABRO1 WT or G338R and analyzed by (L) immunoblot with anti‐Myc or tubulin antibodies, (M) FISH with a telomere 16ptelo probe 4 h after release from a double thymidine block, or (N and O) telomere FISH with a (CCCTAA)3 repeat probe of metaphase spreads. (M) Graphical representation of the frequency of cells with telomere doublets in S phase. Average of two independent experiments (n = 40–58 cells each) ± SEM. *P ≤ 0.05; Student's unpaired t‐test. (N, O) Graphical representation of the frequency of (N) sister telomere loss or (O) fragile telomeres. Average of two independent experiments (n = 632–932 chromosome ends) ± SEM. *P ≤ 0.05; Student's unpaired t‐test.
Source data are available online for this figure.
