Summary
Recent advances in instrumentation and data analysis in field flow fractionation and multi-angle light scattering (FFF-MALS) have enabled greater use of this technique to characterize and quantitate viruses. In this study, the FFF-MALS technique was applied to the characterization and quantitation of type A influenza virus particles to assess its usefulness for vaccine preparation. The use of FFF-MALS for quantitation and measurement of control particles provided data accurate to within 5% of known values, reproducible with a coefficient of variation of 1.9 %. The methods, sensitivity and limit of detection were established by analyzing different volumes of purified virus, which produced a linear regression with fitting value R2 of 0.99. FFF-MALS was further applied to detect and quantitate influenza virus in the supernatant of infected MDCK cells and allantoic fluids of infected eggs. FFF fractograms of the virus present in these different fluids revealed similar distribution of monomeric and oligomeric virions. However, the monomer fraction of cell grown virus has greater size variety. Notably, β-propialactone (BPL) inactivation of influenza viruses did not influence any of the FFF-MALS measurements. Quantitation analysis by FFF-MALS was compared to infectivity assays and real-time RT-PCR (qRT-PCR) and the limitations of each assay were discussed.
Keywords: Influenza virus, Vaccine, Field Flow Fractionation, Multiangle light scattering
1. Introduction
The influenza virus is a globally important respiratory pathogen that causes significant morbidity and mortality in humans and animals (Cox et al., 2004). Vaccination is the primary method for preventing influenza infection and its potentially severe complications. New influenza virus genotypes continuously emerge due to frequent evolutionary events such as genetic re-assortment, recombination and mutation. These events often result in structural changes in the two major influenza virus antigens that induce protective immunity, hemagglutinin (HA) and neuraminidase (NA). Constant changes to viral surface proteins require influenza vaccines to be updated on a yearly basis, to provide protection against contemporary “seasonal” virus strains.
Currently, most seasonal influenza vaccines are trivalent, i.e. it includes three distinct influenza viruses: two influenza A viruses, H3N2 and H1N1 subtypes, and one influenza B virus. Influenza vaccine preparation is a complex process, which involves propagation of viruses in embryonated eggs or cultured cells prior to concentration, inactivation, purification, formulation, and final testing. The majority of clinical isolates do not grow in quantities sufficient to support vaccine production and thus a key aspect of vaccine production is the development of high-growth vaccine viruses. To improve the growth properties of new influenza A viruses in eggs, the production of a reassortant between circulating virus strains and the egg-adapted, laboratory-derived H1N1 strain A/Puerto Rico/8/1934 (PR8), has become standard practice (Kilbourne, 1969). PR8, originally a human isolate, has been passaged extensively in eggs and is considered an attenuated influenza strain in humans (Beare et al., 1975; Neumann et al., 2005). The influenza A viruses used in contemporary vaccine production are high-growth reassortants and usually contain 6 genes (PB2, PB1, PA, NP, M and NS) derived from PR8 and 2 (HA and NA) genes from the presently circulating virus (6:2 re-assortment)(Gerdil, 2003; Neumann et al., 2005).
The World Health Organization (WHO), with its partners, monitors influenza globally and bi-annually recommends the seasonal influenza vaccine composition for both the Northern and Southern Hemisphere influenza seasons. The WHO also coordinates the development and stockpiling of influenza vaccines designed to protect against potential pandemic influenza strains, such as highly pathogenic avian influenza (HPAI) viruses. HPAI vaccine candidate viruses, such as H5N1, are generated using a reverse genetics strategy whereby a low-pathogenic variant of the wild type virus is produced as a 6:2 PR8 reassortant which lacks the multi-basic cleavage site in the HA gene, thus rendering it safer and easier to handle for vaccine production (O’Neill and Donis, 2009). Each of the steps required for development of seasonal and pandemic influenza vaccines, such as production of reassortant viruses, screening for growth characteristics, as well as optimization and adaptation of viruses for growth in eggs or in cell culture require quantitative monitoring to optimize virus yields.
Various techniques are currently used to quantitate viruses in liquid samples. Virus concentration in terms of its infectious dose has classically been determined by measuring plaque forming units (pfu) (Huprikar and Rabinowitz, 1980), 50% tissue culture infective dose (TCID50) (Grigorov et al., 2011), or 50% egg infective dose (EID50) (Huprikar and Rabinowitz, 1980; Reed, 1938). However, these methods are time-consuming and labor-intensive. Other techniques, such as flow cytometry (Schulze-Horsel et al., 2008), enzyme-linked immunosorbent assays (ELISA) (Huaguang, 2006), fluorescent focus assays (FFA) (Wei et al., 2007) and quantitative reverse-transcription polymerase chain reaction (qRT-PCR) (Payungporn et al., 2008) greatly reduce quantitation time but rely on antisera to specific influenza virus antigens or require additional manipulation, such as isolating viral RNA and preparing qRT-PCR standards. The hemagglutination (HA) assay is frequently used for quantitating influenza virus, and is probably the most commonly used assay for comparing viral titers and characterizing virus growth (Kalbfuss et al., 2008; Voeten et al., 1999). The HA assay relies upon HA at the surface of influenza viruses binding to receptors on red blood cells (RBC). However, the haemagglutinating ability of influenza A viruses can vary, depending on the virus subtype and the species of RBC used, leading to inconsistent and inaccurate results amongst different virus strains (McVernon et al., 2010). Therefore, a simple and reliable method that determines influenza virus concentrations, irrespective of strain and subtype, is desirable for quality control of throughput vaccine production.
Asymmetrical field flow fractionation coupled with multi-angle laser light scattering detection (FFF-MALS) was shown to be an efficient approach for size separation and subsequent quantitation of macromolecules and particles (Giddings, 1993), virus-like particles (VLPs) (Lipin et al., 2008; Pease et al., 2009), bacteria (Ping et al., 2007; Polk et al., 2002) and viruses (Saifer, 2001; Wei et al., 2007). It is a rapid and simple technique that enables quantitation without time-consuming sample preparation steps. FFF-MALS combines two analytical methods. FFF is a unique liquid chromatography technique wherein sample separation occurs in a laminar flow channel with no column media to interact with the sample. Particles elute in order of increasing size, and separation is rapid and gentle for the sample. The eluting particles can be detected by MALS, which provides simultaneous detection of light scattered from several angles. By measuring the intensity and angular dependency of the scattered light, it is possible to deduce the radius of the particles and subsequently calculate the number of particles per volume. FFF-MALS was recently applied to characterize influenza virus particles (McEvoy et al., 2011; Wei et al., 2007). Virus particle quantitation by FFF-MALS correlated well with results from other methods, including size exclusion chromatography (SEC-MALS), qRT-PCR, transmission electron microscopy (TEM), and atomic force microscopy (AFM).
In the present study, FFF-MALS analysis was used to quantitate and characterize type A influenza virus, a common and quickly mutating serotype that comprises the majority of vaccine candidates. As most contemporary influenza A vaccine components are derived from a reassortant between PR8 and a circulating influenza virus, the laboratory strain, PR8 was used for these studies. Results show that FFF-MALS can successfully detect and quantitate influenza virions within allantoic fluids of infected chicken eggs and in the supernatant of infected cultured cells. Moreover, it was demonstrated that FFF-MALS can quantitate purified influenza viruses treated with β-propialactone (BPL), which is used for virus inactivation in early stages of vaccine preparation (Perrin and Morgeaux, 1995). Finally, a comparison of the total number of virus particles quantitated by FFF-MALS to that detected by EID50, virus plaque assay, and quantitative real-time RT-PCR (qRT-PCR) was performed. The usefulness and limitations of each assay is discussed.
2. Materials and Methods
2.1. Viruses and cell lines
The influenza virus A/Puerto Rico/8/1934 (H1N1) (PR8) and the reassortant candidate vaccine virus A/Indonesia/05/2005(H5N1)/PR8-IBCDC-RG2 carrying the HA and NA from A/Indonesia/5/05 (H5N1) and internal genes from A/PR/8/34 strains were propagated in 10-day-old embryonated chicken eggs at 34 °C for 48 h, after which the allantoic fluid containing the virus was harvested. Allantoic fluids containing H5N1 reassortant virus were treated with BPL for 48 h at 4 °C (Perrin and Morgeaux, 1995). Allantoic fluids containing PR8 were divided into two; one half of the fluid was treated with BPL and the other half was left untreated. Virus-containing fluids then were clarified by low speed centrifugation at 1,000 xg for 20 min. Virus inactivation was confirmed by inoculation tests over two passages in embryonated chicken eggs. Both untreated and BPL-treated allantoic fluids were then concentrated by ultracentrifugation at 30,000 xg for 3 h, and purified by sucrose density centrifugation (Liu et al., 2002) in BLS-3 facilities. Virus stocks were then stored at −80 °C prior to use.
Madin–Darby canine kidney (MDCK) cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and were maintained in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10 % fetal bovine serum, 0.1 % penicillin/streptomycin, and 0.1 % glutamine. Confluent cell monolayers were washed twice with phosphate-buffered saline (PBS; Gibco, Carlsbad, CA, USA) and infected with 0.001 multiplicity of infection (MOI) of influenza PR8. After incubation for 1 h at 37 °C, the cells were washed and the medium was replaced with DMEM supplemented with 0.1 % penicillin/streptomycin, 0.1 % glutamine, and 2 μg/ml L-(tosylamido-2-phenyl) ethyl chloromethyl ketone (TPCK) trypsin. After 48 h incubation, the supernatant was harvested, clarified by low speed centrifugation as described above, and used for quantitative analysis.
2.2. Plaque assay
The plaque assay was used to measure the amount of infectious virus particles in a sample, as described previously (Huprikar and Rabinowitz, 1980). Briefly, MDCK cells in 6-well culture plates (1x106 cells/well) were infected with virus samples serially diluted in DMEM. Duplicate 0.1 ml aliquots of serial 10-fold dilutions were added to each well. After infection for 1 hour at 37 °C, the wells were washed twice with DMEM, and 3 ml overlay medium was added to each well. The overlay medium consisted of DMEM with 0.1 % penicillin/streptomycin, 0.1 % glutamine, 0.08 % of SeaKem LE Agarose (Lonza, Basel, Switzerland), and 1 μg/μl -TPCK trypsin. The plates were placed in 5 % CO2 at 37 °C for 72 h for plaque development. After the incubation period, the soft agar overlay was decanted gently, and 1 ml of 4 % formaldehyde-water solution containing 0.1 % crystal violet was added per well. After 30 min at room temperature, the plates were washed with water and visible plaques were counted.
2.3. Hemagglutination (HA) assay
Virus-containing fluids (50 μl) were mixed with the same volume of 0.5 % turkey RBC (tRBC) in PBS in V-bottom microtiter plates. Agglutination of the tRBC was determined after 30 – 60 min incubation at room temperature (World Health Organization, 2011).
2.4. Measurement of 50 % egg infectious dose (EID50)
The EID50 of influenza virus was determined as previously described (Huprikar and Rabinowitz, 1980). Briefly, serial 10-fold dilutions of the virus were prepared in PBS, and 100 μl of each dilution was inoculated into the chorio-allantoic cavities of 10-days-old embryonated chicken eggs. The eggs were incubated at 34 °C for 48 h. Four eggs were infected with each virus dilution. Harvested allantoic fluid was tested for HA activity using 0.5 % tRBC.
2.5 One-step qRT- PCR
The M gene-specific primers and probe set was designed for the detection of all type A influenza viruses, comprising M-For (5′-CATGGAATGGCTAAAGACAAGACC-3′), M-Rev (5′-AGGGCATTTTGGACAAAGCGTCTA-3′) and M-Probe (FAM-ACGCTCACCGTGCCCAGT-BHQ1). M gene DNA from a PR8 influenza A virus was previously cloned into the pAMP1 vector and amplified by PCR with a M gene forward primer (5′-GATCGCTCTTCAGGGAGCAAAAGCAGGTAG-3′) and the reverse primer with a T7 promoter added to the 5′ end (T7/M-Rev; 5′-TGTAATACGACTCACTATAGGGCATTT TGGACAAAGCGTC-3′). The resulting PCR product was then used for in vitro transcription of M gene RNA using the Riboprobe in vitro Transcription Systems, according to the manufacturer (Promega, Madison, WI, USA). Following DNase treatment and RNA extraction, the concentration of RNA was measured by spectrophotometry and converted to copy number/μl. Dilutions of 1010 copies/μl of standard M gene RNA were prepared and stored at −80 °C until further use. Influenza virus RNA was extracted in duplicate using the Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA, USA) and eluted in a final volume of 30 μL of RNase-free water. 5 μl of RNA was used for qRT-PCR in a single-step reaction using the Qiagen QuantiTect Probe RT-PCR Kit. Each reaction was run in parallel with standard concentrations of M gene RNA (1010–102 copies/μl) to produce a standard curve used for interpolation of sample viral RNA copy number. qRT-PCR was performed according to the manufacturer’s protocol with 35 cycles using the Mx3005P QPCR system (Stratagene, La Jolla, CA, USA). Cycling conditions included an initial reverse transcription step at 50 °C for 30 min, then an initial denaturation step at 95 °C for 15 min, and 35 amplification cycles of denaturation at 94 °C for 15 sec, followed by final annealing/extension at 60 °C for 30 sec.
2.6. Transmission electron microscopy (TEM)
For negative stain electron microscopy preparation, virus grown in embryonated chicken eggs was concentrated and mixed 1:1 with 4 % phosphate-buffered formaldehyde. Copper mesh grids coated with formvar and carbon were placed on a drop of the specimen for 10 min, dried, and stained with 2 % phosphotungstic acid, pH 7.3. Samples were examined on an FEI Tecnai Spirit transmission electron microscope (Hillsboro, OR, USA). Images were recorded digitally as TIF files using a bottom-mounted AMT camera (Woburn, MA, USA).
2.7. FFF-MALS
Asymmetric field flow fractionation (FFF) was performed using an Agilent auto-sampler and pump (Agilent, Santa Clara, CA, USA) connected to a Wyatt Eclipse-3 AFFF separation module (Wyatt, Santa Barbara, CA, USA), and a Wyatt DAWN HELEOS 18-angle MALS detector. The MALS system was equipped with a gallium-arsenic laser (658 nm) and measurements were obtained at 25 °C by detectors situated at angles of 44°, 50°, 57°, 64°, 72°, 81°, 90°, 99°, 108°, 117°, 126°, 134° and 144° to the incident beam. Separations were performed using a 10 kDa molecular weight cut-off polyethersulfone (PES) membrane in a 27 cm (long) separation channel with a 350 μm spacer, according to the methods of Wei et al. (2007). Based on the size of influenza virus particles, between 80–130 nm in diameter (Stanley, 1944), the system was validated using polystyrene microspheres with nominal diameters of 100 nm and 200 nm (Polysciences, Warrington, PA, USA). The radii measured by FFF-MALS for both microspheres (93.4 ± 0.6 nm and 45.5 ± 0.5 nm) were within the coefficient of variation (CV) values (4.6 % and 5.2 % for 200 nm and 100 nm microspheres, respectively) reported in the manufacturer’s certificates of analyses.
Samples were introduced to the channel at 0.2 ml/min and subsequently focused at the head of the channel at a focus flow rate of 0.4 ml/min. Samples were eluted over 35 min with a channel flow rate of 1 ml/min and a cross flow gradient of 0.3–0 ml/min. Viral particles were detected with the MALS detector and the radius of gyration (RMS radius) of the eluted species was calculated by the method of Wyatt et al. (1993), using Astra V software. To quantitate the number of particles, the virion shape was substituted for the sphere model to calculate a geometric radius for particles. The total number of virus particles, as well as the relative numbers of monomers and aggregates, was calculated using ASTRA® software as described (Wyatt, 2004), assuming a refractive index of 1.59 for polystyrene particles and 1.5 for influenza virions.
Experimental buffer (EB, 0.01 M sodium/potassium phosphate pH 7.4 containing 0.14 M sodium chloride) was used to prepare and run all samples. Polystyrene microsphere standards (Polysciences, Warrington, PA, USA) were prepared and analyzed in EB with an additional 0.08 % (w/v) sodium dodecyl sulphate to reduce bead aggregation. For virus analysis, allantoic fluids were diluted 10 times with EB, infected cell supernatants were used without dilution, while purified influenza virus was diluted with EB to a final viral protein concentration of 5 μg/ml, and 100 μl of the final solution was used for analysis.
2.7. Limits of Detection
Detection limit, the smallest amount or concentration of a particular substance that can be reliably detected in a given sample, was calculated according to Gomez-Taylor et al. (U.S. Environmental Protection Agency. 2003). The limit of detection (LOD) is defined as 3 standard deviations from the control sample reading, while the limit of quantitation (LOQ) is defined as 10 standard deviations from the control. PBS was used as a control sample to establish LOD and LOQ; mean and standard deviation were calculated from seven different measurements of PBS. The average 90° light scattering signal of PBS was estimated as 15.6 mV, with standard deviation ±0.04 mV. The calculated LOD and LOQ were 15.7 mV and 16.0 mV, respectively. The virus samples with light scattering signals greater than 16.0 mV were selected for number-particles quantitation analysis. The method detection limit (MDL) was calculated by the Hubaux-Vos procedure (Hubaux and Vos, 1970). MDL is defined as the minimum concentration of substance that can be measured and reported with 99 % confidence.
3. Results
3.1. Fractionation and characterization of influenza virus particles with FFF-MALS
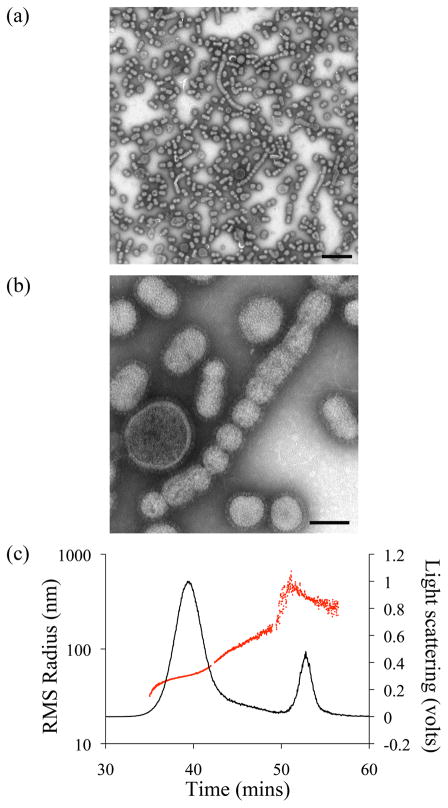
The suspension of purified PR8 influenza virus propagated in embryonated eggs contained a mixture of individual virions of different sizes and morphology, as well as aggregates (Fig. 1a and b). The majority of individual virions had spherical or ovoid morphology. The average diameter of spherical virions estimated by TEM was 104 ± 30 nm (n=100), whereas virions with ovoid morphology had average dimensions of 145 x 88 ± 30 nm with an average geometrical diameter of 116 ±30 nm (n=100).
Figure 1.
Analysis of purified Influenza PR8 virus by TEM (a and b) and FFF-MALS (c). TEM images show purified virions with different sizes and morphologies at (a) low magnification (scale bar = 500 nm) and (b) high magnification (scale bar = 100 nm), respectively. The average diameter of spherical virions estimated by TEM is 104 nm (radius – 52 nm). The average size of ovoid virions is 145 x 88 nm (radius 72 x 44nm). (c) The FFF-MALS fractogram of the virus shows the 90° light scattering signal (black line) during 31 to 61 min overlaid with RMS radius (red line).
The virion sizes estimated by TEM were comparable to those detected by FFF-MALS. Individual components of purified influenza PR8 virus were fractionated by FFF and characterized with MALS for particle size and scattering intensity. Figure 1c shows a typical FFF-MALS fractogram of purified influenza PR8 virus. The main peak eluted between 35 and 42 min and contained virus particles with an RMS radius of 30–70 nm. The first peak overlapped with that of the larger particles (RMS 70–150 nm) appearing as a “shoulder” on the fractogram. Another major peak, eluted at 49–56 min, represented particles with an RMS radius of 150–500 nm (Fig. 1c). The experimentally measured RMS radius of gyration was converted to geometrical radius based on the sphere model, as incorporated within the ASTRA® software package. The calculated average geometrical diameter of virion species in the first peak was 112.6 ± 8 nm, corresponding to monomeric virions. The “shoulder” peak included particles with average geometrical diameter of 170 ± 8 nm and likely represented filamentous virions and small aggregates, while the second peak contained virus aggregates with a calculated average geometrical diameter of 270 ± 8 nm. The number of monomers was calculated using data for particles with an RMS radius of 30–70 nm, while all particles with an RMS radius over 70 nm were assumed to be aggregates. To assess assay reproducibility, three samples of equal concentration were analyzed independently on different days (Table 1). Quantitative analyses were accurately reproducible, with a coefficient of variation (CV) of 1.9 %. Approximately 92 % of influenza virions from the purified virus population were monomeric, and only 8 % were present as aggregates (Table 1).
Table 1.
Reproducibility of total number virions counts and percent of aggregates detected by FFF-MALS.
| Experiment | Injection | Virions per injection (Log10) | % Monomers | % Aggregates |
|---|---|---|---|---|
| 1 | 1 | 8.5 | 94 | 6 |
| 2 | 8.4 | 93 | 7 | |
|
| ||||
| 2 | 1 | 8.1 | 91 | 9 |
| 2 | 8.2 | 92 | 8 | |
| 3 | 8.1 | 91 | 9 | |
|
| ||||
| 3 | 1 | 8.1 | 91 | 9 |
| 2 | 8.1 | 91 | 9 | |
| 3 | 8.1 | 93 | 7 | |
|
| ||||
| Average | 8.2 | 92 | 8 | |
| % CV | 1.9 | 1.3 | 15 | |
3.2. Detection limit and method sensitivity
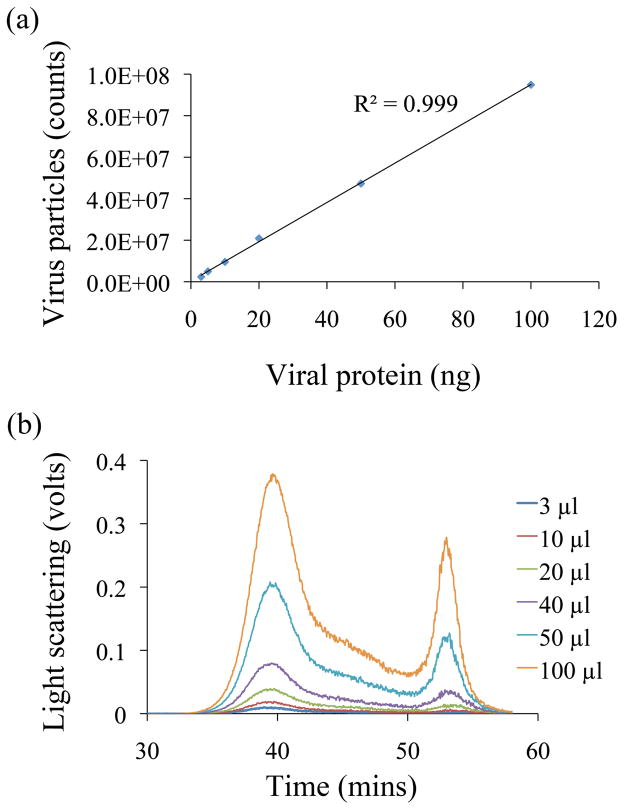
The FFF-MALS detection limit for influenza virus particles was evaluated by injecting different volumes (3–100 μL) of purified PR8 (2 μg/ml). Figure 2 demonstrates the linear regression between the total number of virus particles counted and injected volume (R2 = 0.99) within a 1-log10 range of total virus particle counts from 2.37 x 106 to 9.56 x 107. The MDL estimated from this slope (Hubaux and Vos) was 4.3 x 106 particles. LOD and LOQ estimated using the slope by extrapolating to zero were 2.2 x 106 and 2.3 x 106 virus particles, respectively.
Figure 2.
FFF-MALS detection limits for purified influenza A/PR/8/34. (a) Positive slope of total virus particles counts derived from the light scattering data. (b) FFF-MALS fractograms of the purified virus injected at increasing volumes (3, 5, 10, 20, 50 and 100 μl).
The method sensitivity was comparable to that of the HA assay, and the number of virus particles estimated by FFF-MALS was compared to the HA titer (Table 2). In experiments, the lowest level of detection of purified influenza PR8 by FFF-MALS was 2.35 x 106, which corresponded to 4 HA units.
Table 2.
Correlation of total number particles counts estimated by FFF-MALS, and HA titer of influenza A/PR/8/34.
| Injected volume (μL) | HA titer | Virus particles (x106) |
|---|---|---|
| 100 | 128 | 95.6 |
| 50 | 64 | 47.8 |
| 20 | 32 | 20.9 |
| 10 | 16 | 10.5 |
| 5 | 8 | 5.2 |
| 3 | 4 | 2.37 |
3.3. Detection of influenza virus in various samples
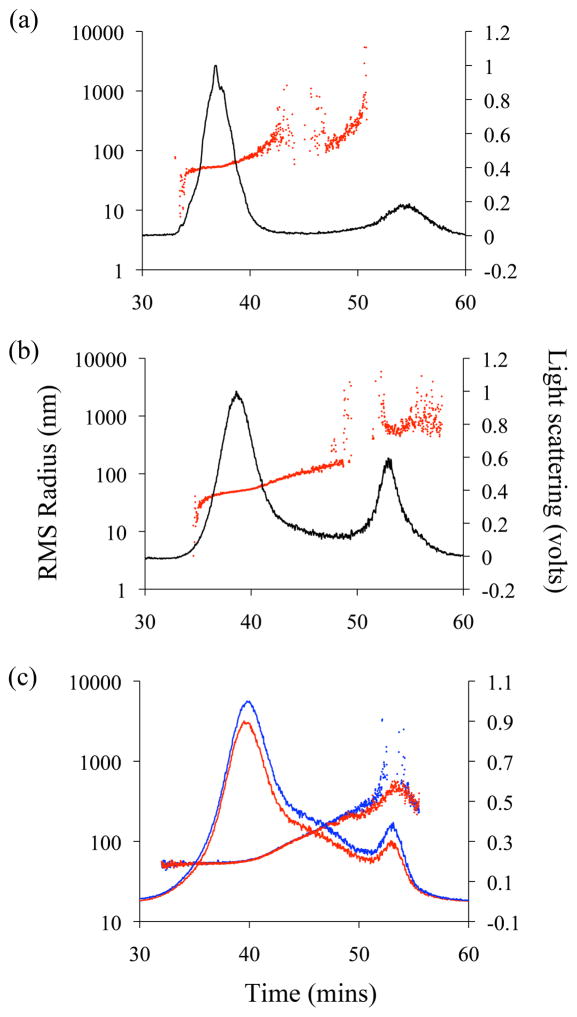
We applied FFF-MALS to directly detect and quantitate influenza virus in the allantoic fluids of infected embryonated chicken eggs or in supernatant of infected MDCK cells. 10-day-old embryonated eggs or confluent MDCK cells were infected with influenza PR8, allantoic fluid and supernatants were collected 48 h after infection, clarified, and subjected to fractionation and quantitation by FFF-MALS. Figure 3 represents a typical FFF fractogram of virus-containing supernatant of infected MDCK cells (Figure 3a) and allantoic fluids collected from infected eggs (Figure 3b). The major influenza PR8 monomer-containing fraction in both fluids eluted between 34–42 min, and a second peak, representing large aggregates, eluted between 49–60 min. The monomer-containing fraction from MDCK cell supernatants contained viruses with an RMS radius of 30–100 nm (average of geometrical diameter 122 ± 6 nm), while virus monomers from allantoic fluid ranged from 30–70 nm (average of geometrical diameter 112 ± 1.2 nm). The percentage of aggregation was extremely low in both the supernatant and allantoic fluid samples (2 % and 3 %, respectively; Table 3).
Figure 3.
FFF-MALS analysis of the influenza virus in various samples. FFF-MALS fractogram of the supernatant of infected MDCK cells (a), allantoic fluids from infected eggs (b), and purified untreated (blue) and BPL-treated (red) influenza A/PR/8/34 (c). The 90° light scattering signal over time is overlaid with RMS radius.
Table 3.
Comparison of total virus counts and relative percent of aggregates percent in different species
| PR/8/34 virus source | Experiments | Injections | Particles (Log10)/injection | Monomers (%) | Aggregates (%) |
|---|---|---|---|---|---|
| Purified | 2 | 6 | 8.1 ± 0.04 | 92 ± 0.8 | 8 ± 0.8 |
| Purified, BPL treated | 2 | 6 | 8.1 ± 0.09 | 92 ± 1.4 | 8 ± 1.4 |
| Allantoic fluids of infected eggs | 2 | 6 | 8.1 ± 0.12 | 97 ± 1 | 3 ± 1 |
| Supernatant from infected MDCK cells | 3 | 7 | 8.5 ± 0.08 | 98 ± 0.6 | 2 ± 0.6 |
We applied FFF-MALS to investigate whether BPL inactivation affected the biophysical (i.e., size distribution and monomer-aggregate percentages) and quantitative characteristics of influenza viruses. Figure 3c illustrates FFF-MALS fractograms of PR8, treated or not treated with BPL, respectively. These results confirmed that BPL inactivation affected neither the biophysical nor quantitative characteristics of the virus (Table 3).
3.4. Comparison of total virus particle numbers analyzed by different methods
Four different influenza PR8 virus samples were quantitated by EID50, PFU, qRT-PCR, and FFF-MALS. These samples comprised purified viruses treated or not treated with BPL, allantoic fluid of infected embryonated chicken eggs, and supernatant of infected MDCK cells (Table 5). Each sample was evaluated in three independent experiments, and the mean and standard deviation of these results calculated. Comparison of the quantitative analyses showed the number of influenza virus particles detected by FFF-MALS in the purified virus samples was approximately 10-fold higher than those detected by EID50 and PFU. However, the number of M gene copies detected by qRT-PCR for these samples was about 10-fold greater than that estimated by FFF-MALS.
4. Discussion
Influenza viruses cause significant morbidity and mortality, resulting in large numbers of hospitalizations each year. Vaccination against influenza continues to be the primary strategy for preventing disease (Cox et al., 2004). Production of influenza virus vaccines has traditionally relied on virus grown in embryonated hen eggs. However, cell lines have also been established as a substrate for vaccine virus production and vaccines produced in these systems were shown to have equivalent efficacy to those grown in eggs (Bardiya and Bae, 2005). The process of influenza vaccine manufacturing includes multiple steps. Initially, preparation of the vaccine strain and optimization of virus growth conditions occurs before vaccine bulk manufacture. To reduce risks from handling and improve growth, WHO laboratories generate a 6:2 reassortant candidate vaccine virus, containing 6 internal virus genes derived from PR8 and 2 genes HA and NA from presently circulating virus (Kilbourne, 1969; Neumann et al., 2005). Vaccine manufacturers use this hybrid to optimize growth conditions for both egg- and cell-based vaccines (World Health Organization, 2009). A robust quantitative and qualitative method for monitoring influenza viruses in allantoic fluids and infected cell culture supernatants can greatly increase the speed and efficiency of vaccine production.
Although live attenuated influenza vaccines are available, most influenza vaccines produced globally are inactivated, and BPL is commonly used for the inactivation process (Bardiya and Bae, 2005; Kistner et al., 1998; Liu et al., 2002). Virus inactivation is an important component of influenza vaccine production. Inactivation requires that the virus is rendered non-infectious whilst preserving the integrity of immunogenic epitopes, and this is currently achieved using chemical treatment with formalin or BPL (World Health Organization, 2006). Importantly, BPL treatment does not alter the antigenic properties of inactivated virions and BPL-inactivated virus retains the ability to elicit a protective immune response. However, viruses inactivated by treatment with BPL are unable to replicate (Hovden et al., 2005) and cannot be quantitated by biological methods such as plaque assay or EID50 of TCID50 and rely only on remaining haemagglutination activity. Additional information on virion integrity as well as their aggregation state would be very desirable especially for production of influenza vaccine based on whole virus particles.
FFF-MALS analysis has previously been shown to be effective in separating and quantitating purified or unpurified concentrated influenza type A and B virus (Wei et al., 2007). In the present study, this technique was further applied to quantitate type A influenza viruses in solution, and focused on two early steps of vaccine preparation; firstly, detection and quantitation of virus harvested from eggs and cell cultures and, secondly, virus quantitation following inactivation.
Using PR8, the influenza A strain used to generate high-growth reassortants for current influenza vaccines, quantitation by FFF-MALS showed linearity with increasing particle numbers (R2 = 0.99) and reproducibility of the assay correlated well between experiments (CV = 1.9 %). By TEM, the average size of particles within the monomer fraction of egg grown viruses varied in geometrical diameter from 104 ±30 nm for spherical to 116 ±30 nm for ovoid virions, respectively, and correlated well with FFF-MALS calculations (112.6 ±8 nm). These data, together with previous studies involving influenza viruses (Wei et al., 2007), validate the application of FFF-MALS for the quantitation of influenza viruses. Comparative analyses of influenza viruses present in the supernatant of infected cells and egg allantoic fluid revealed similar FFF elution profiles, with 97–98 % of viruses from both sources eluted in the monomer fraction. However, the RMS radius of viruses in the monomer-containing fraction from cultured cell supernatants were more variable, ranging from 30 to 100 nm (average geometrical diameter 122 ±6 nm), as compared to the 30–70 nm range (average geometrical diameter 112.6 ±8 nm) for egg grown viruses. These elevated values for virus radii from infected MDCK cells could be explained by an increased production of filamentous virions, which has previously been shown in influenza viruses propagated from polarized epithelial cells, including MDCK cells (Roberts and Compans, 1998).
FFF-MALS was applied to quantitate influenza virus following treatment with BPL and assess the influence of BPL treatment upon virion size and aggregation. Data showed that the biophysical and quantitative characteristics of viruses treated with BPL are equivalent to untreated viruses, and confirm that BPL treatment does not affect the measurement of influenza viruses by FFF-MALS. Quantitative analysis results by FFF-MALS were compared to those obtained by other standard methods, including PFU, EID50, qRT-PCR and HA. Each type of assay measured distinct attributes of virions, resulting in variations in the measured quantity of influenza virus. Quantitation of virus by PFU and EID50 was one log10 lower than the amount of virions detected by FFF-MALS, because PFU and EID50 detected only infectious virions, while both infectious and non-infectious virions are present in virus stocks (Donald and Isaacs, 1954; Nayak et al., 2009). This result is consistent with a previous determination that approximately ten virus particles correspond to one EID50 (Donald and Isaacs, 1954). qRT-PCR is an indirect measurement technique, often used for quantitative analysis of virus through measurement of the number of viral gene copies. This method relies on the amplification of the viral gene and on the standard serial dilution. In this study, the number of matrix (M) gene copies detected by qRT-PCR was one log10 higher than the number of viral particles estimated by FFF-MALS. However, there is evidence that virus samples contain virus particle-associated nuclease-resistant RNA, which can artificially increase the number gene copies (Dovas et al., 2010; Wei et al., 2007). It is also possible that FFF-MALS quantitation may underestimate the number of virus particles counted, due to increased error when virus is eluted within the aggregate fraction (McEvoy et al., 2011).
The hemagglutination assay (HA), one of the most widely used assays for indirect quantitation of influenza viruses (Mahy, 1996), is commonly applied to compare virus titers and characterize virus growth (Kistner et al., 1998; Tree et al., 2001; Voeten et al., 1999). However, the hemagglutinating ability of influenza A viruses varies for each influenza subtype, the host system used for virus propagation, and the species of the RBC used for the assay. For example, 2009 pandemic H1N1 virus agglutinates chicken, human, guinea pig and turkey RBC equally well, while recent H3N2 viruses typically agglutinate guinea pig RBC better than others, and H5N1 viruses agglutinate horse RBC better than others (McEvoy et al., 2011). Thus, several types of RBCs may often be needed for the HA assay within a single study, complicating the analyses. In this study, virus detection by FFF-MALS, was at a similar level of sensitivity to the HA assay. The lowest virus concentration detected with FFF-MALS was 2.35 x 106 particles (7.8 x 105 per μl), which was in the quantitative range (LOQ - 2.3 x 106) and corresponded to 4 HA units (Table 2). Taken together, this data demonstrates that FFF-MALS is a reliable technique for the detection and characterization of influenza viruses with sensitivity and reproducibility of data that is at least equivalent to currently available laboratory assays.
In comparison with traditional virus quantitation methods such as EID50 or PFU analysis, which take several days, or qRT-PCR which requires labor-intensive preparation of the RNA and standards, FFF-MALS allows the user to obtain results for each sample within one hour and eliminates many sample preparation steps. The assay is automated and allows continuous consecutive analysis of approximately 100 samples. FFF-MALS can directly quantify influenza virus in supernatants of infected cells or in allantoic fluid of infected eggs with minimal sample preparation (clarification by low speed centrifugation) and is, therefore, highly time-effective. Although influenza virus quantitation by FFF-MALS cannot substitute for other assays that measure the biological activity of a virus, it can provide useful information on virus populations in solution. Using FFF-MALS to screen the biophysical growth characteristics of reassortant viruses can provide critical information on the number, size distribution and aggregation state of vaccine candidate virus when grown under different conditions. Thus, FFF-MALS analysis in collaboration with another biological assay could be a useful technique to assess virus growth and yield during influenza vaccine optimization and downstream production.
Table 4.
Comparison of total number virus particles counted by different methods
| PR/8/34 virus | Experiment | EID50 (Log10) | PFU (Log10) | qRT-PCR (Log10) | FFF- MALS (Log10) |
|---|---|---|---|---|---|
| Purified (5 μg/ml) | 1 | 7.3 | 7 | 9.4 | 8.2 |
| 2 | 7.5 | 7.2 | 9.3 | 8.2 | |
| 3 | 7.3 | 7.6 | 8.9 | 8.4 | |
| Mean ± SD | 7.4 ± 0.1 | 7.3 ± 0.3 | 9.2 ± 0.3 | 8.3 ± 0.1 | |
|
| |||||
| Allantoic fluids of infected eggs (1:10 dilution with PBS) | 1 | 7.1 | 7.2 | 8.9 | 8.1 |
| 2 | 7.1 | 7.4 | 8.9 | 8.1 | |
| 3 | 7.3 | 7.2 | 9.9 | 8.3 | |
| Mean ± SD | 7.2 ± 0.1 | 7.3 ± 0.1 | 9.2 ± 0.6 | 8.2 ± 0.1 | |
|
| |||||
| Supernatant from infected MDCK cells (undiluted) | 1 | 7.5 | 7.3 | 10.3 | 8.5 |
| 2 | 7.5 | 7.1 | 9.8 | 8.6 | |
| 3 | 7.8 | 7.4 | 9.6 | 8.7 | |
| Mean ± SD | 7.6 ± 0.2 | 7.3 ± 0.2 | 9.9 ± 0.4 | 8.6 ± 0.1 | |
|
| |||||
| Purified, BPL treated (5 μg/ml) | 1 | N/Aa | N/A | 8.5 | 8.2 |
| 2 | N/A | N/A | 8.7 | 8.3 | |
| 3 | N/A | N/A | 8.9 | 8.3 | |
| Mean ± SD | 8.7 ± 0.2 | 8.3 ± 0.06 | |||
|
| |||||
| Purified, BPL treated RGA/Indonezia/5/05 (1.5 μg/ml) | 1 | N/A | N/A | 8.1 | 7.4 |
| 2 | N/A | N/A | 8.5 | 7.3 | |
| 3 | N/A | N/A | 8.1 | 7.2 | |
| Mean ± SD | 8.2 ± 0.2 | 7.3 ± 0.1 | |||
N/A-Not applicable
Acknowledgments
This work was funded by the Centers for Disease Control and Prevention. We would like to thank Michelle Chen and the technical scientists at the Wyatt Technology Corporation for their careful reading of the manuscript and helpful advice. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry.
References
- Bardiya N, Bae JH. Influenza vaccines: recent advances in production technologies. Appl Microbiol Biotechnol. 2005;67:299–305. doi: 10.1007/s00253-004-1874-1. [DOI] [PubMed] [Google Scholar]
- Beare AS, Schild GC, Craig JW. Trials in man with live recombinants made from A/PR/8/34 (H0N1) and wild H3N2 influenza viruses. Lancet. 1975;2:729–732. doi: 10.1016/s0140-6736(75)90720-5. [DOI] [PubMed] [Google Scholar]
- Cox RJ, Brokstad KA, Ogra P. Influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand J Immunol. 2004;59:1–15. doi: 10.1111/j.0300-9475.2004.01382.x. [DOI] [PubMed] [Google Scholar]
- Donald HB, Isaacs A. Counts of influenza virus particles. J Gen Microbiol. 1954;10:457–464. doi: 10.1099/00221287-10-3-457. [DOI] [PubMed] [Google Scholar]
- Dovas CI, Papanastassopoulou M, Georgiadis MP, Chatzinasiou E, Maliogka VI, Georgiades GK. Detection and quantification of infectious avian influenza A (H5N1) virus in environmental water by using real-time reverse transcription-PCR. Appl Environ Microbiol. 2010;76:2165–2174. doi: 10.1128/AEM.01929-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdil C. The annual production cycle for influenza vaccine. Vaccine. 2003;21:1776–9. doi: 10.1016/s0264-410x(03)00071-9. [DOI] [PubMed] [Google Scholar]
- Giddings JC. Field-flow fractionation: analysis of macromolecular, colloidal, and particulate materials. Science. 1993;260:1456–1465. doi: 10.1126/science.8502990. [DOI] [PubMed] [Google Scholar]
- Grigorov B, Rabilloud J, Lawrence P, Gerlier D. Rapid titration of measles and other viruses: optimization with determination of replication cycle length. PLoS One. 2011;6:e24135. doi: 10.1371/journal.pone.0024135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovden AO, Cox RJ, Madhun A, Haaheim LR. Two doses of parenterally administered split influenza virus vaccine elicited high serum IgG concentrations which effectively limited viral shedding upon challenge in mice. Scand J Immunol. 2005;62:342–352. doi: 10.1111/j.1365-3083.2005.01666.x. [DOI] [PubMed] [Google Scholar]
- Huaguang L. Dot-ELISA for the Detection of Avian Influenza Virus. 2006/0246429 A1 Patent # US. 2006
- Hubaux A, Vos G. Decision and detection limits for linear calibration curves. Anal Chem. 1970;42:849–855. [Google Scholar]
- Huprikar J, Rabinowitz S. A simplified plaque assay for influenza viruses in Madin-Darby kidney (MDCK) cells. J Virol Methods. 1980;1:117–120. doi: 10.1016/0166-0934(80)90020-8. [DOI] [PubMed] [Google Scholar]
- Kalbfuss B, Knochlein A, Krober T, Reichl U. Monitoring influenza virus content in vaccine production: precise assays for the quantitation of hemagglutination and neuraminidase activity. Biologicals. 2008;36:145–161. doi: 10.1016/j.biologicals.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Kilbourne ED. Future influenza vaccines and the use of genetic recombinants. Bull World Health Organ. 1969;41:643–645. [PMC free article] [PubMed] [Google Scholar]
- Kistner O, Barrett PN, Mundt W, Reiter M, Schober-Bendixen S, Dorner F. Development of a mammalian cell (Vero) derived candidate influenza virus vaccine. Vaccine. 1998;16:960–968. doi: 10.1016/s0264-410x(97)00301-0. [DOI] [PubMed] [Google Scholar]
- Lipin DI, Chuan YP, Lua LH, Middelberg AP. Encapsulation of DNA and non-viral protein changes the structure of murine polyomavirus virus-like particles. Arch Virol. 2008;153:2027–2039. doi: 10.1007/s00705-008-0220-9. [DOI] [PubMed] [Google Scholar]
- Liu T, Muller J, Ye Z. Association of influenza virus matrix protein with ribonucleoproteins may control viral growth and morphology. Virology. 2002;304:89–96. doi: 10.1006/viro.2002.1669. [DOI] [PubMed] [Google Scholar]
- McEvoy M, Razinkov V, Wei Z, Casas-Finet JR, Tous GI, Schenerman MA. Improved particle counting and size distribution determination of aggregated virus populations by asymmetric flow field-flow fractionation and multiangle light scattering techniques. Biotechnol Prog. 2011;27:547–554. doi: 10.1002/btpr.499. [DOI] [PubMed] [Google Scholar]
- McVernon J, Laurie K, Nolan T, Owen R, Irving D, Capper H, Hyland C, Faddy H, Carolan L, Barr I, Kelso A. Seroprevalence of 2009 pandemic influenza A(H1N1) virus in Australian blood donors, October–December 2009. Euro Surveill. 2010;15(40) doi: 10.2807/ese.15.40.19678-en. pii=19678. [DOI] [PubMed] [Google Scholar]
- Nayak DP, Balogun RA, Yamada H, Zhou ZH, Barman S. Influenza virus morphogenesis and budding. Virus Res. 2009;143:147–161. doi: 10.1016/j.virusres.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Fujii K, Kino Y, Kawaoka Y. An improved reverse genetics system for influenza A virus generation and its implications for vaccine production. Proc Natl Acad Sci USA. 2005;102:16825–16829. doi: 10.1073/pnas.0505587102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill E, Donis RO. Generation and characterization of candidate vaccine viruses for prepandemic influenza vaccines. Curr Top Microbiol Immunol. 2009;333:83–108. doi: 10.1007/978-3-540-92165-3_4. [DOI] [PubMed] [Google Scholar]
- Payungporn S, Crawford PC, Kouo TS, Chen LM, Pompey J, Castleman WL, Dubovi EJ, Katz JM, Donis RO. Influenza A virus (H3N8) in dogs with respiratory disease, Florida. Emerg Infect Dis. 2008;14:902–908. doi: 10.3201/eid1406.071270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease LF, 3rd, Lipin DI, Tsai DH, Zachariah MR, Lua LH, Tarlov MJ, Middelberg AP. Quantitative characterization of virus-like particles by asymmetrical flow field flow fractionation, electrospray differential mobility analysis, and transmission electron microscopy. Biotechnol Bioeng. 2009;102:845–855. doi: 10.1002/bit.22085. [DOI] [PubMed] [Google Scholar]
- Perrin P, Morgeaux S. Inactivation of DNA by beta-propiolactone. Biologicals. 1995;23:207–211. doi: 10.1006/biol.1995.0034. [DOI] [PubMed] [Google Scholar]
- Ping L, Buchler R, Mithofer A, Svatos A, Spiteller D, Dettner K, Gmeiner S, Piel J, Schlott B, Boland W. A novel Dps-type protein from insect gut bacteria catalyses hydrolysis and synthesis of N-acyl amino acids. Environ Microbiol. 2007;9:1572–1583. doi: 10.1111/j.1462-2920.2007.01279.x. [DOI] [PubMed] [Google Scholar]
- Polk HC, Jr, Bowden TA, Jr, Rikkers LF, Balch CM, Murie JA, Pories WJ, Buechler MH, Neoptolemos JP, Fazio VW, Schwartz SI, Cameron JL, Kelly KA, Grosfeld JL, McFadden DW, Souba WW, Pruitt BA, Jr, Johnston KW, Rutherford RB, Arrequi ME, Scott-Conner CE, Warshaw AL, Sarr MG, Cuschieri A, MacFadyen BV, Thompkins RK. Scientific data from clinical trials: investigators’ responsibilities and rights. Dis Colon Rectum. 2002;45:725–726. doi: 10.1007/s10350-004-6285-y. [DOI] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Am J Epidemiol. 1938;27:493–497. [Google Scholar]
- Roberts PC, Compans RW. Host cell dependence of viral morphology. Proc Natl Acad Sci U S A. 1998;95:5746–5751. doi: 10.1073/pnas.95.10.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saifer MGP, Williams D. Quantitation of viruses by Light Scattering. Patent # US. 2001;6:316, 185 B1. [Google Scholar]
- Schulze-Horsel J, Genzel Y, Reichl U. Flow cytometric monitoring of influenza A virus infection in MDCK cells during vaccine production. BMC Biotechnol. 2008;8:45. doi: 10.1186/1472-6750-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley WM. The Size of Influenza Virus. J Exp Med. 1944;79:267–83. doi: 10.1084/jem.79.3.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tree JA, Richardson C, Fooks AR, Clegg JC, Looby D. Comparison of large-scale mammalian cell culture systems with egg culture for the production of influenza virus A vaccine strains. Vaccine. 2001;19:3444–3450. doi: 10.1016/s0264-410x(01)00053-6. [DOI] [PubMed] [Google Scholar]
- U.S. Environmental Protection Agency. Technical support document for the assessment of detection and quantitation approaches. 2003. EPA-HQ-OW-2003-0002-0110. [Google Scholar]
- Voeten JT, Brands R, Palache AM, van Scharrenburg GJ, Rimmelzwaan GF, Osterhaus AD, Claas EC. Characterization of high-growth reassortant influenza A viruses generated in MDCK cells cultured in serum-free medium. Vaccine. 1999;17:1942–1950. doi: 10.1016/s0264-410x(98)00464-2. [DOI] [PubMed] [Google Scholar]
- Wei Z, McEvoy M, Razinkov V, Polozova A, Li E, Casas-Finet J, Tous GI, Balu P, Pan AA, Mehta H, Schenerman MA. Biophysical characterization of influenza virus subpopulations using field flow fractionation and multiangle light scattering: correlation of particle counts, size distribution and infectivity. J Virol Methods. 2007;144:122–132. doi: 10.1016/j.jviromet.2007.04.008. [DOI] [PubMed] [Google Scholar]
- World Health Organization. [last accessed 2nd July 2013];A review of production technologies for influenza virus vaccines, and their suitability for deployment in developing countries for influenza pandemic preparedness. 2006 http://who.int/vaccine_research/diseases/influenza/Flu_vacc_manuf_tech_report.pdf.
- World Health Organization. [(last accessed: 2nd July 2013)];Pandemic influenza vaccine manufacturing process and timeline. 2009 http://www.who.int/csr/disease/swineflu/notes/h1n1_vaccine_20090806/en/
- World Health Organization. Manual for the laboratory diagnosis and virological surveillance of influenza. World Health Organization Press; Geneva: 2011. [Google Scholar]
- Wyatt PJ. Light scattering and the absolute characterization of macromolecules. Analytica Chimica Acta. 1993;272:1–40. [Google Scholar]
- Wyatt PJ, Weida MJ. Method and apparatus for determining absolute number densities of particles in suspension. Patent # US. 2004;6:774, 994 B1. [Google Scholar]