Figure 6.
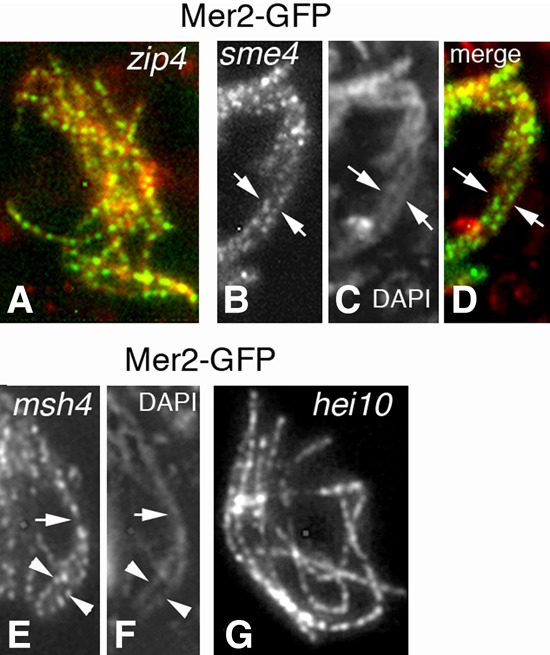
Mer2 localization in the SC plus recombination mutants. (A) In absence of Zip4, Mer2 foci are regularly spaced along all unsynapsed axes like in wild type (distances of 0.42 µm ± 0.06 µm) from leptotene through late pachytene (by ascus size). (B–D) The same timing occurs in the absence of Sme4. As this mutant is not required for homolog pairing, Mer2 foci decorate both coaligned homologs (arrows). (C) Corresponding DAPI. (D) Merge. (E,F) In the absence of Msh4, SCs form only partially: Mer2 remains on axes in unsynapsed regions (arrowheads) but shifts to SC central regions in segments with formed SCs (arrow). (F) Corresponding DAPI. (G) In hei10-null pachytene nuclei, Mer2 localizes along all SCs like in wild type and with similar spacing. Bars, 2 µm.
