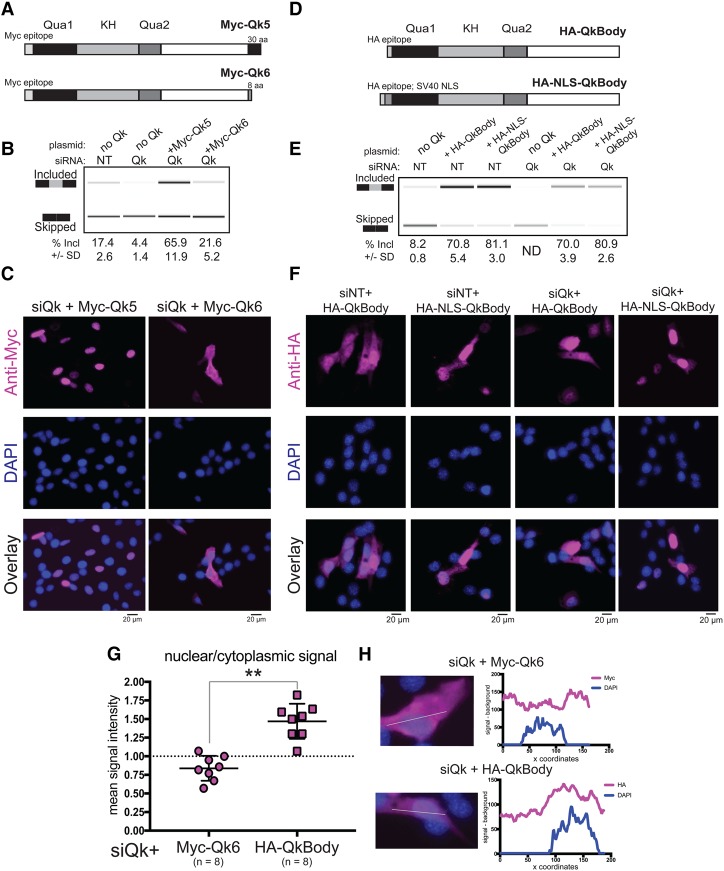
Figure 2.
Nuclear localization is sufficient for Qk splicing function. (A) Model of Myc epitope-tagged Qk5 and Qk6 with the number of unique amino acids at the C terminus shown. (B, top) RT–PCR products from Dup-Capzb exon 9 splicing reporter analyzed on a Bioanalyzer from RNA extracted from C2C12 myoblasts transfected with either control siRNA (siNT) or siRNA targeting all Qk isoforms (siQk) or cotransfected with both siQk and Myc-Qk5 or siQk and Myc-Qk6. The mean percentage included is shown below with standard deviation from three independent biological replicates. (C) Indirect immunofluorescence using anti-Myc epitope antibody (magenta; top), DAPI (blue; middle), and channels overlaid (bottom) of C2C12 myoblasts transfected with siQk + Myc-Qk5 (left) or siQk + Myc-Qk6 (right). Bar, 20 µm. (D) Model of Qk mutant proteins tested for splicing function and localization: HA-QkBody consists of an N-terminal HA epitope tag and the 311 amino acids common to all Qk proteins (top), and HA-NLS-QkBody consists of an N-terminal HA tag and nuclear localization signal (NLS) from SV40 preceding the 311 amino acids common to all Qk isoforms (bottom). (E) RT–PCR products from Capzb exon 9 splicing reporter analyzed on a Bioanalyzer from RNA extracted from C2C12 myoblasts transfected with (from left to right) siNT, siNT and HA-QkBody, siNT and HA-NLS-QkBody, siQk, siQk and HA-QkBody, or siQk and HA-NLS-QkBody. The mean percentage included from three independent biological replicates with standard deviation is shown below. (F) Indirect immunofluorescence using HA antibody (magenta; top), DAPI staining (blue; middle), and overlaid images (bottom) of representative C2C12 myoblasts from (from left to right) siNT + HA-QkBody, siNT + HA-NLS-QkBody, siQk + HA-QkBody, and siQk + HA-NLS-QkBody transfections. Bar, 20 µm. (G) Graph showing nuclear/cytoplasmic signal intensity of Myc-Qk6 (left) or HA-QkBody (right) staining of C2C12 myoblasts transfected with siRNA targeting all Qk isoforms. n = 8 siQk. (**) P ≤ 0.01. (H, left) Representative images from which epitope tag and DAPI signal intensity were measured for data shown in G. Plots at the right show corresponding signal intensities (minus background; Y-axis) plotted over image coordinates (X-axis) based on the line drawn through representative images at the left.