FIG 1.
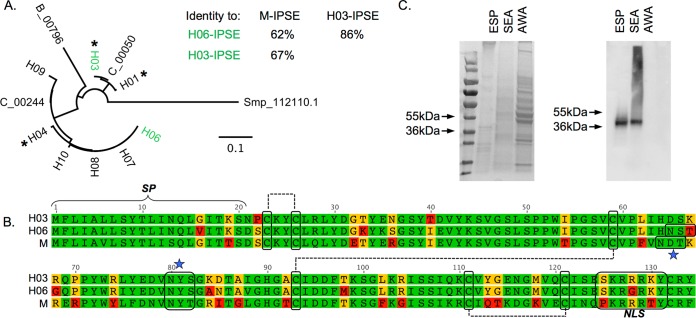
S. haematobium expresses multiple forms of H-IPSE. (A) Amino acid sequences of predicted H-IPSE paralogs, M-IPSE, and sequenced transcripts from egg cDNA were globally aligned by using a Blosum62 cost matrix, and a tree was built by using the neighbor-joining method in Geneious 7.1.4. The bar represents amino acid substitutions per site. H06 and H03 variants chosen for expression are highlighted in green, and their respective amino acid identities and identities to M-IPSE are shown. All variants identified through 3′-RACE cloning are indicated with asterisks. (B) Alignment of amino acid sequences of H03-IPSE (top rows) and H06-IPSE (middle row)s with the sequence of the homolog in S. mansoni (IPSE/alpha-1; here named M-IPSE) (bottom rows). These H-IPSE clones retain ∼63 to 68% amino acid identity and several features described previously for M-IPSE (24), including a 20-amino-acid classical signal sequence, seven cysteine residues involved in disulfide bonds, two N-linked glycosylation consensus motifs, and a predicted nuclear localization sequence. Residues shown in green are identical, residues in yellow share properties (e.g., hydrophobicity or polarity), and residues in red lack similarity. (C) SDS-PAGE gel (left) and Western blot with anti-H06-IPSE antiserum (right). Lanes contain parasite-derived adult worm antigen (AWA), egg secretory protein (ESP), or soluble egg antigen (SEA).