FIG 1.
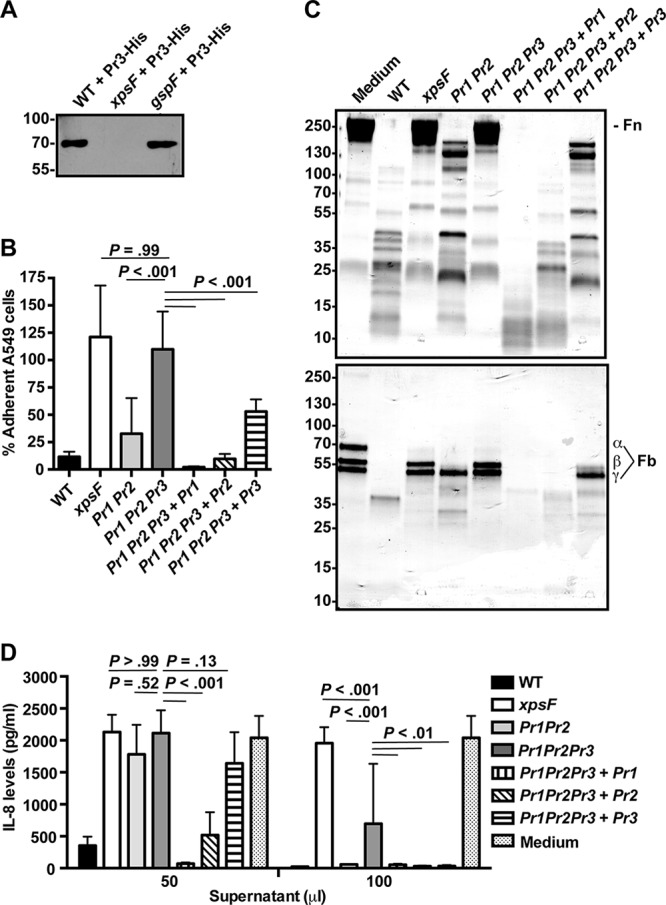
Contribution of StmPr3 to Xps-mediated A549 cell detachment and Xps-mediated degradative activities. (A) His-tagged StmPr3 (Pr3-His) was exogenously produced from a plasmid in strain K279a (WT), xpsF mutant NUS4 (xpsF), and gspF mutant NUS1 (gspF). Supernatants collected from the cultures of these strains were analyzed by SDS-PAGE, followed by immunoblotting for anti-His. The migration of molecular mass standards (in kilodaltons) is indicated to the left of the gel images. (B) A549 cells were incubated for 24 h with 25% (vol/vol) culture supernatant collected from the WT strain, xpsF mutant NUS4, stmPr1 stmPr2 mutant NUS7 (Pr1 Pr2), stmPr1 stmPr2 stmPr3 mutant NUS14 (Pr1 Pr2 Pr3), or the stmPr1 stmPr2 stmPr3 mutant complemented with stmPr1 (Pr1 Pr2 Pr3 + Pr1), stmPr2 (Pr1 Pr2 Pr3 + Pr2), or stmPr3 (Pr1 Pr2 Pr3 + Pr3). Cell detachment was determined by quantifying the remaining adherent cells. Data were normalized to those for cells treated with medium alone, for which adherence was set at 100%. (C) Ten micrograms of human fibronectin (Fn; top) or fibrinogen (Fb; bottom) was incubated at 37°C for 16 h with 25 μl of culture supernatant from the WT, the xpsF mutant, the indicated protease mutant and complemented strains, or a control treated with medium alone. ECM protein degradation was analyzed by SDS-PAGE and total protein staining with Coomassie. The fibrinogen α, β, and γ chains are indicated. The migration of molecular mass standards (in kilodaltons) is indicated to the left of the gel images. Data are representative of those from three independent experiments. (D) Recombinant IL-8 was incubated at 37°C for 16 h with 50 or 100 μl of culture supernatant collected from the WT strain, the xpsF mutant, the indicated protease mutant and complemented strains, or a control treated with medium alone. IL-8 levels were quantified by ELISA. For panels B and D, the data are represented as the mean and SD from three independent experiments.
