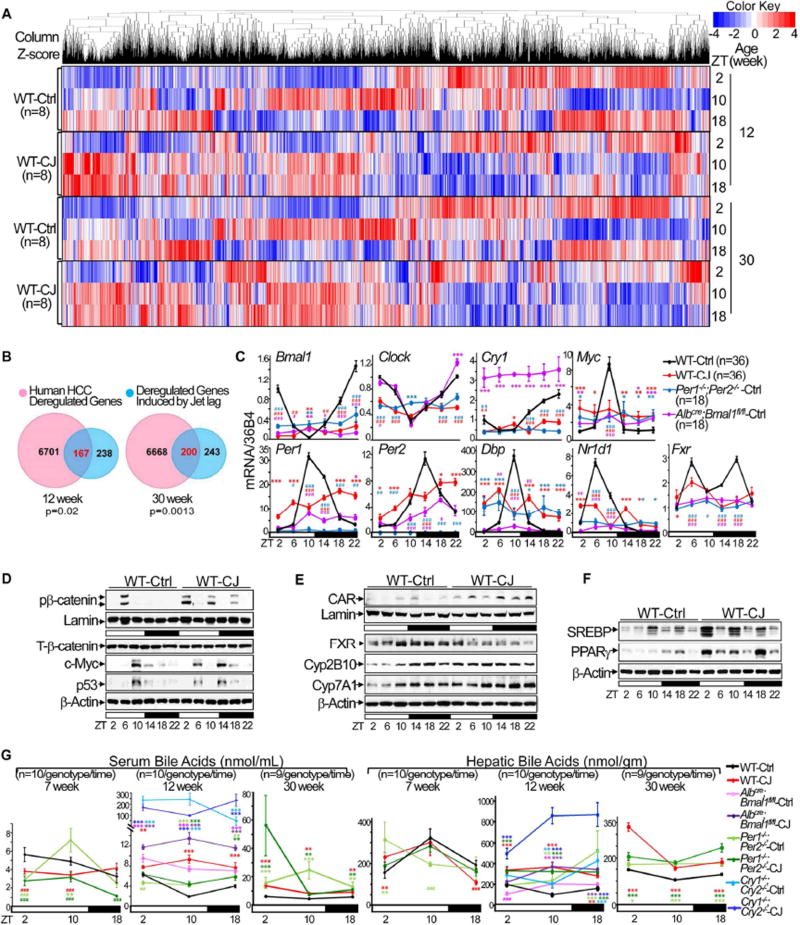
Figure 4. Chronic jet lag induces genome-wide gene deregulation in mouse livers.
(A) Hierarchical clustering heat map shows expression of hepatic genes in the livers of control and jet-lagged WT mice. (B) Venn diagrams showing persistent overlap of deregulated liver transcriptomic signatures in jet-lagged WT mice with that of human HCC from 12 to 30 weeks of age (hypergeometric test). (C) The summary of 3 independent RT-PCR studies on the expression of core circadian genes, Myc and Fxr in the livers of 12 week-old circadian gene mutant and WT mice, with levels detected in control WT mice at ZT2 as the arbitrary unit 1. (D–F) Western blots show the expression of Ser552 phospho-β-catenin (pβ-catenin), total β-catenin (T-β-catenin), c-Myc and p53 in (D), FXR, CAR, Cyp2B10 and Cyp7A1 in (E), and SREBP1 and PPARγ in (F) in the livers of 12 week old WT mice. (G) Serum and hepatic bile acid levels over a 24 hr period in circadian gene-mutant and WT mice at 7, 12 and 30 weeks of age. *: Increase,#: decrease, comparing to WT controls at the same time, */#p<0.05, **/##p<0.01; ***/###p<0.001, student t test, ±SEM. See also figure S6 and Table S1–6.