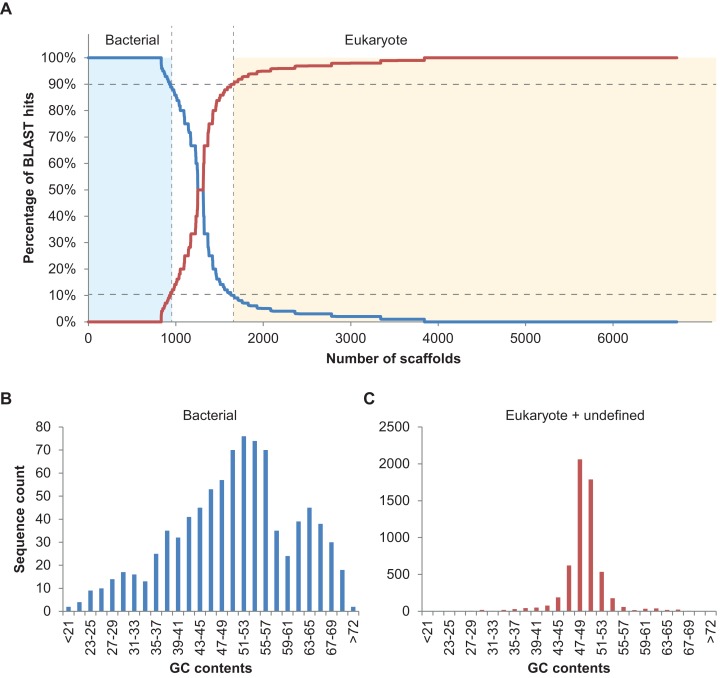
Figure 2. Taxoblast analysis of the S. japonica genome.
Application of the Taxoblast pipeline to identify potential bacterial sequences in the published S. japonica genome (Ye et al., 2015). (A) shows the percentage of bacterial/eukaryote blast hits over the 6,731 scaffolds >2 kbp with blast hits (254 scaffolds >2kbp had no hits). Dotted lines show the 90% cutoff proposed to consider a sequence as “contaminant”. (B) and (C) illustrate the different distribution of GC contents in the sequences considered bacterial, and those considered eukaryotic or unclassified.