Abstract
Context
Oropharyngeal squamous cell carcinoma (OPSCC) is associated both with tobacco use and human papillomavirus (HPV) infection. It is argued that carcinogen-driven tumorigenesis is a distinct disease from its virally-driven counterpart. We hypothesized that tumorigenesis is the result of a loss of genotypic robustness resulting in an increase in phenotypic variation in tumors compared to adjacent histologically normal tissues, and that carcinogen-driven tumorigenesis results in greater variation than their virally-driven counterparts.
Objective
To examine the loss of robustness in carcinogen- and virally-driven OPSCC samples, and to identify potential pathways involved.
Design
We used coefficients of variation (CVs) for mRNA and microRNA expression to measure the loss of robustness in OPSCC samples. Tumors were compared to matched normal tissues, and were further categorized by HPV and patient smoking status. Weighted gene co-expression networks were constructed for genes with highly variable expression among the HPV- tumors from smokers.
Results
We observed more genes with variable mRNA expression in tumors compared to normal tissues, regardless of HPV and smoking status, and more microRNAs with variable expression in HPV− and HPV+ tumors from smoking patients than from non-smokers. For both the mRNA and microRNA data, we observed more variance among HPV− tumors from smokers compared to HPV+ tumors from non-smokers. The gene co-expression network construction highlighted pathways that have lost robustness in carcinogen-induced tumors but appear stable in virally-induced tumors.
Conclusions
Using CVs and co-expression networks, we identified multiple altered pathways that may play a role in carcinogen-driven tumorigenesis.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the 7th most common cancer worldwide1. Risk factors include tobacco use, alcohol consumption, and Human Papillomavirus (HPV) infection (predominantly HPV type 16)2–4. HPV+ tumors are frequently found within the oropharynx3. Several studies have previously demonstrated that HPV− and HPV+ tumors are clinically and molecularly distinct diseases5. Molecular differences between HPV− and HPV+ tumors include differential gene expression patterns6,7, methylation patterns8,9, and microRNA (miRNA) levels10. Additionally, patient response to treatment is dramatically improved for HPV+ tumors11,12, and disease specific survival is increased compared to HPV− patients13–15. These essential differences may be due to different mechanisms of tumorigenesis. Since both virally-induced and carcinogen-induced tumors exist within the oropharynx, oropharyngeal squamous cell carcinoma (OPSCC) is an appropriate model to study how these two mechanisms of tumorigenesis differ in genetic variation. We hypothesize that one of the underlying mechanisms of HNSCC is the loss of cellular integrity, specifically the loss of robustness.
Robustness is a phenomenon whereby environmental as well as the underlying genetic variation does not get expressed at the phenotypic level16. We now know that this phenomenon may be the result of multiple factors, however the original assumption by Waddington was that given a trait under selection, the mechanism that brings about this trait, i.e. the gene regulatory network that results in the phenotype under observation, would be under selection17–19. Since variation due to random mutation is an unavoidable process, Waddington argued that any mechanism that would buffer such variation to be expressed at the phenotypic level would be selected for. Theoretical studies by Bergman and Siegal show that this is not a necessary condition for the evolution of robustness 20,21. One way to achieve robustness is to have hierarchical and complex genotype-phenotype maps22. Complex traits, namely traits that result from complex genotype-phenotype maps, may exhibit robustness even though there may not be direct selection for a particular phenotype. This phenomenon of robustness allows the harboring of underlying genetic variation that is considered cryptic23,24.
The genotype is fairly easy to describe: it is the specific sequence for the genetic material for all biological organisms. The phenotype is less obvious. In this study we look at gene expression as a fundamental level of the phenotype. Thus, the phenomenon of robustness that we are examining is the insensitivity of the gene expression level, our phenotype, to genetic variation. Robustness may seem to hinder evolution, however, at times of stress, when a major perturbation occurs, cryptic variation percolates to the phenotypic level enabling selection. Such perturbation may be the result of major environmental changes or large genotypic aberration. Cancer, we argue, may be the outcome resulting from such breakage of robustness.
In contrast to breaking of robustness in carcinogen-driven tumorigenesis, viral infections, such as HPV, Hepatitis B virus, and Epstein-Barr virus, induce tumorigenesis through systematic and potentially predictable mechanisms 25–28. Viruses essentially highjack the infected cell’s molecular machinery to promote their own survival. In the process, the virus needs to shut down apoptotic and cellular senescence signals, which can ultimately lead to tumorigenesis. Since the virus uses the same mechanisms for shutting down tumor suppressive pathways in each infected cell, variation at the level of gene expression is hypothesized to be lower among multiple patient tumors.
Unlike previous studies of tumorigenesis and progression, we look at cancer as the result of normal, non-tumorigenic cells losing the robustness of their complex genotype-phenotype maps. As a result, we hypothesized that tumors will exhibit a greater degree of phenotypic variation, as measured by gene expression, than their histologically normal counterparts. To study the increase in expression variation, i.e. loss of robustness, within OPSCC, we designed an approach to detect genes that exhibit a high degree of variability in expression across tumor samples. Our approach is not looking at the consequences of consistent up and down regulation for genes involving tumor suppressors or oncogenes, but looks at variation that is increased in pathways. From our results, we identified multiple genes involved in cancer pathways that have previously been associated with cancer, as well as singled out those not yet directly implicated in tumorigenesis. We tested the hypothesis that variability in expression will differ in environmentally-driven cancers (tobacco smoking) vs. virally-driven cancers (HPV infection). Patient tumors were compared to adjacent normal tissues, which are considered to be “wild-type” thus robust, while the tumor cells are considered to be those in which robustness is broken. In turn, we were able to apply our method to HPV- tumors from smoking patients and HPV+ tumors non-smoking patients to identify genes that are highly variable within the context of smoking. Finally, we also applied the same approach to microRNA expression data to assess the loss of robustness at an epigenetic level.
Materials and Methods
Patient Tissue Samples
Histologically confirmed OPSCC tumors were obtained by biopsy or surgical resection from patients undergoing treatment at Montefiore Medical Center in Bronx, NY. All patients provided written consent for participation in this study under a protocol approved by the Institutional Review Board at Montefiore Medical Center. Tumors and adjacent histologically normal tissue were snap-frozen in liquid nitrogen within 30 minutes of surgical resection or biopsy. Samples were kept at −80°C until used for microarray analyses.
All tumors were tested for HPV DNA and RNA (including for HPV-16, the most prevalent type found in OPSCC) using Polymerase Chain Reaction (PCR) assays as described previously9. One sample was negative for HPV-16 but positive for HPV-35. HPV type 35 is closely related phylogenetically to HPV-1629. Tumors testing positive for low-risk (non-oncogenic) types were considered HPV−. The sample sizes used to analyze the mRNA and microRNA datasets are listed in Table 1.
Table 1.
Sample Sizes.
| HPV− Smokers | |
|---|---|
| mRNA Array | |
| Number of Tumors | 20 |
| Number of Tumors with Matched Normal Tissue | 15 |
| miRNA Array | |
| Number of Tumors with Matched Normal Tissue | 14 |
| HPV− Non-Smokers | |
|---|---|
| mRNA Array | |
| Number of Tumors | 4 |
| Number of Tumors with Matched Normal Tissue | 4 |
| HPV+ Smokers | |
|---|---|
| mRNA Array | |
| Number of Tumors | 21 |
| Number of Tumors with Matched Normal Tissue | 16 |
| miRNA Array | |
| Number of Tumors with Matched Normal Tissue | 18 |
| HPV+ Non-Smokers | |
|---|---|
| mRNA Array | |
| Number of Tumors | 6 |
| Number of Tumors with Matched Normal Tissue | 4 |
| miRNA Array | |
| Number of Tumors with Matched Normal Tissue | 3 |
There were not enough HPV− tumors from non-smokers to do a CV analysis for the miRNA array.
CV= Coefficient of Variation
HPV= Human Papillomavirus
miRNA= microRNA
mRNA= messenger RNA
mRNA Microarray Preparation
The protocol for the miRNA expression microarrays was previously described9. For each primary tumor and matched normal sample, total RNA (500 ng) was amplified and biotin labeled with the Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX). Whole genome expression was analyzed by RNA hybridization to the Illimina HumanHT-12-v3 Expression BeadChip (Illumina, San Diego, CA). Probes were matched to known genes and alternative splice variants using the RefSeq database release 17 and UniGene build 188. Controls for each RNA sample were used to confirm RNA quality, biotin labeling success, hybridization stringency, and signal levels. 39,364 probes from the microarray were used for subsequent analyses.
Microarray expression values were quantile normalized within BeadStudio (Illumina, San Diego, CA) prior to analysis. Expression data was batch corrected using the ComBat function from the sva R package30. For each sample, the median expression values from 750 negative control genes were subtracted from the other expression values, and expression values ≤ 1 were converted to a value of 1. Only genes with 70% of their values > 1 were considered valid and included in subsequent analyses. Table 2 lists the final number of probes analyzed for each group.
Table 2.
Percentage of Probes Used for mRNA Analyses.
| Group | Number of Probes | % of Total Probes |
|---|---|---|
|
| ||
| HPV− Smokers (T-N) | 15,670 | 40% |
| HPV− Non-Smokers (T-N) | 16,784 | 43% |
| HPV+ Smokers (T-N) | 16,167 | 41% |
| HPV+ Non-Smokers (T-N) | 16,700 | 42% |
| HPV− Smokers and HPV+ Non-Smokers | 14,285 | 36% |
There were a total of 39,364 probes used for the mRNA analyses (not including controls).
HPV= Human Papillomavirus
mRNA= messenger RNA
N= Normal
T= Tumor
Coefficients of Variation for mRNA Data
Coefficients of variation (CV, σ/μ) were calculated for each gene with valid expression across 70% of the samples. CVs were plotted to detect differences in genetic robustness between tumor and normal tissues. The differentiation of CV(tumor)–(normal) and CV(normal)–(tumor) were used to identify potentially relevant genes in HPV− and HPV+ tumor-normal pairs. The threshold for high CV values is case dependent but has been reported to be as high as 0.5 for gene expression studies31. We increase the threshold of the CV value for genes with highly variable expression to 1 as to ensure that the expression levels of the genes selected were indeed highly variable. Then, the CV of the genes were plotted for 20 HPV− and 6 HPV+ tumor samples. A threshold of CV(HPV−) > 1 and CV(HPV+) < 0.4 was used to identify 43 genes with expression levels that were only highly variable in HPV− samples (see Supplemental Figure 1 for threshold selection). To validate the relevancy of these 43 genes an expression heatmap was generated using the supraHex R package32 (www.r-project.org). Patients were clustered according to a dendrogram at the top of the heatmap using a Spearman correlation.
Weighted Gene Co-expression Network Analysis for mRNA Data
The WGCNA (Weighted Gene Co-expression Network Analysis) R package33 was used to establish a co-expression network for selected genes. A typical workflow for WGCNA was used with a few modifications33,34. Briefly, for the 43 selected genes, we used a power adjacency matrix (β=6) to generate modules of co-expressed genes as described by Zhang and Horvath34. The selected genes were placed into four modules based on the Topological Overlap Matrix-based dissimilarity (DistTOM) of their expression. A DistTOM heatmap and multi-dimensional scaling plot were generated to determine which modules would form networks of co-expression. Co-expression networks were visualized in VisANT35 with a weight cutoff of 0.4 for the blue network and 0.5 for the red network. Gene ontology (GO) was then done using PANTHER36 and EMBL-EBI QuickGO (http://www.ebi.ac.uk/QuickGO/, accessed on 1/12/2015).
microRNA Microarray Preparation
The miRNA microarray protocol was previously described37. For each primary tumor and matched normal sample, total RNA (200 ng) was used for miRNA microarray analysis using the Illumina DASL (cDNA-mediated Annealing, Selection, Extension, and Ligation) in the 96-well-plate Sentrix Array Matrix (SAM) format (Illumina, San Diego, CA). RNA quality was assessed using the Agilent Bioanalyzer (Agilent, Santa Clara, CA). Results were analyzed within Illumina’s GenomeStudio (Illumina, San Diego, CA) using their miRNA labeling and hybridization quality controls. Seven hundred and thirty nine miRNAs that satisfied quality control (QC) entries from the microarray were used for subsequent analyses.
Coefficients of Variation for miRNA Data
CV were calculated for each miRNA on the array. Like the mRNA data, CV were plotted for tumor-normal pairs and for HPV− and HPV+ tumors. Thresholds of CV(tumor) – CV(normal) > 1 and CV(normal) – CV(tumor) > 1 were used to determine the number of miRNAs highly variable expression levels in each tumor-normal set. A threshold of CV(HPV−) > 1 and CV(HPV+) < 0.5 was used to select 18 miRNAs with expression levels that were only highly variable in the HPV− tumors. A literature search was performed for the selected miRNAs and those with published data were included in the results. TargetScan38 and Target Explorer (Ingenuity, Redwood City, CA) were used to generate a table of cellular functions for the remaining miRNAs based on their inferred targets.
Results
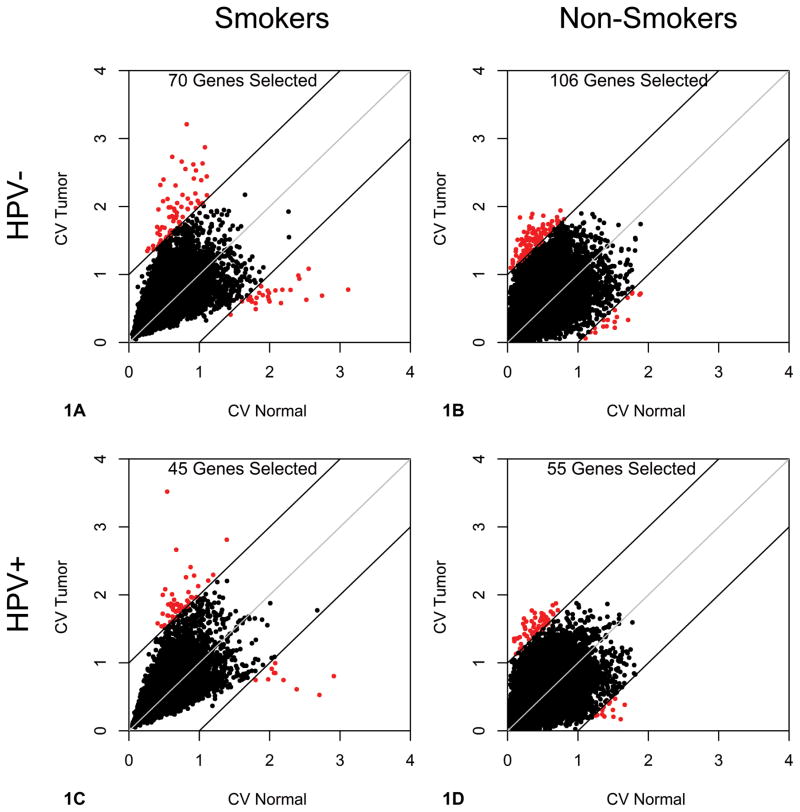
To measure a loss of robustness in OPSCC samples, we examined gene expression. mRNA microarrays provided a high throughput mechanism for measuring gene expression across both HPV− and HPV+ patient samples from the oropharynx. To test our hypothesis that there is a greater number of genes with high expression variance in tumors than in normal tissue, we analyzed the CVs for mRNA expression in HPV− and HPV+ primary tumor-normal pairs (see Materials and Methods). Patients were categorized by HPV and smoking status, resulting in four groups for analysis. Here we define non-smokers as patients who have never smoked tobacco. To visualize expression variation between tumor and normal tissues, the CVs for each gene were plotted, and genes with higher CVs in either tumor or normal tissues were identified (Figure 1A–1D, highlighted in red). The two patient smoking groups had similar sample sizes, with 15 tumor-normal pairs for the HPV− patient samples and 16 pairs for HPV+ patient samples. Likewise, the two non-smoking groups had identical sample sizes, 4 tumor-normal pairs for both HPV− and HPV+ patient samples. From our results, we observe a higher number of genes with variant expression in HPV− tumors compared to the matched normal tissue, 70 vs. 27 genes for smokers (Figure 1A) and 106 vs. 8 genes for non-smokers (Figure 1B). Similarly, there were more genes with highly variable expression in HPV+ tumors than normal tissues, 45 vs. 10 genes for smokers (Figure 1C) and 55 vs. 14 genes for non-smokers (Figure 1D). This confirmed our hypothesis that tumors exhibit more genes with a higher variance in expression than their matched histologically normal tissue. Interestingly, the HPV− and HPV+ tumors from the smoking groups had a larger range of CV values with a maximal CV value above 3, while the maximal CV value for the non-smoking groups did not exceed 2. This translates to a higher degree of expression variation for tumors from smoking patients. Additionally, there were about twice the number of genes with highly variable expression in HPV− tumors compared to HPV+ tumors regardless of smoking status (70 vs. 45 for the smoking patients, and 106 vs. 55 for the non-smoking patients), which supports our second hypothesis that virally-driven tumorigenesis is more controlled than carcinogen-driven tumorigenesis which is induced by random gene mutation.
Figure 1. Tumor messenger RNA (mRNA) Expression Variation in Human Papillomavirus negative (HPV−) and Human Papillomavirus positive (HPV+) Patients.
Scatterplots representing individual mRNA coefficients of variation (CV) for tumors and matched normal tissue. Genes with a CV > 1 both for tumors and for normal tissues are colored red. A. CVs plotted for 15 paired HPV− tumors from smokers. Seventy genes had greater CVs in tumor samples compared to 27 genes in the corresponding normal tissues, suggesting a greater amount of genes with high expression variation among tumors. B. CVs plotted for 4 paired HPV− tumors from non-smokers. Again, there is a greater amount of genes with high expression variation among the tumors, as demonstrated by 106 variably expressed genes in the tumors and only 8 variably expressed genes in the normal tissue. C. CVs plotted for 16 paired HPV+ tumors from smokers. There were 45 variably expressed genes in the tumors compared to 10 variably expressed genes in the normal tissue. D. CVs plotted for 4 paired HPV+ tumors from non-smokers. Similarly, 55 genes were variant in the tumors compared to 14 genes in the matched normal tissue.
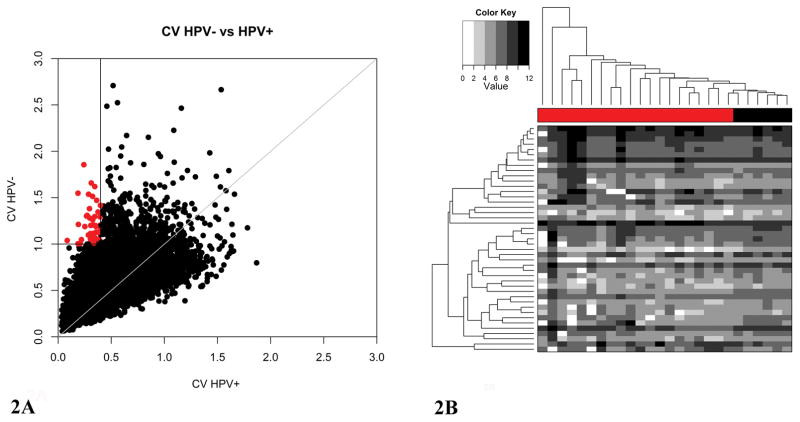
To identify genes with highly variable expression in smoking induced tumors only, the CVs of each gene were plotted for tumors from 20 HPV− smoker and 6 HPV+ non-smoker OPSCC patients (Figure 2A). Since smoking added expression variation to the HPV+ samples, the HPV+ smoking patients were excluded from this analysis. Although the sample sizes between the two groups differed largely (20 vs. 6), as we have shown with the HPV− non-smokers (n=4), the sample sizes did not affect the number of genes with highly variable expression in the tumor samples (Figure 1). Since we define a CV of 1 as high expression variation, CVs greater than 1 in HPV− tumors from smokers and less than 0.4 in HPV+ tumors from non-smokers were considered to be highly variable in smokers only (see Supplemental Figure 1 for HPV+ threshold selection). From this analysis, we identified 43 genes that showed high expression variation only in tumors from HPV− smokers. Expression values for the 43 genes with highly variable expression were then used to group tumor samples by hierarchical clustering (Figure 2B). The HPV+ samples (colored black) formed one cluster at the right end of the dendrogram, while the HPV− samples (colored red) were broken up into several clusters.
Figure 2. Greater Degree of messenger RNA (mRNA) Expression Variation in Human Papillomavirus negative (HPV−) Tumors from Smokers Compared to Human Papillomavirus positive (HPV+) tumors from Non-Smokers.
A. Scatterplot representing individual mRNA coefficients of variation for 20 HPV− tumors from smokers and 6 HPV+ tumors from non-smokers. Forty-three genes with high expression variation only in HPV− tumors are colored red. B. Heatmap representing the expression of the 43 genes with high expression variation in HPV− tumors. The patient sample dendrogram is above the heatmap. The 20 HPV− tumors from smokers are denoted with a red bar, and the 6 HPV+ tumors from non-smokers are indicated by the black bar. The expression values were Log2 transformed for the heatmap, and the grayscale was divided into 6 distinct colors to highlight the extreme differences in expression within the two groups of tumors.
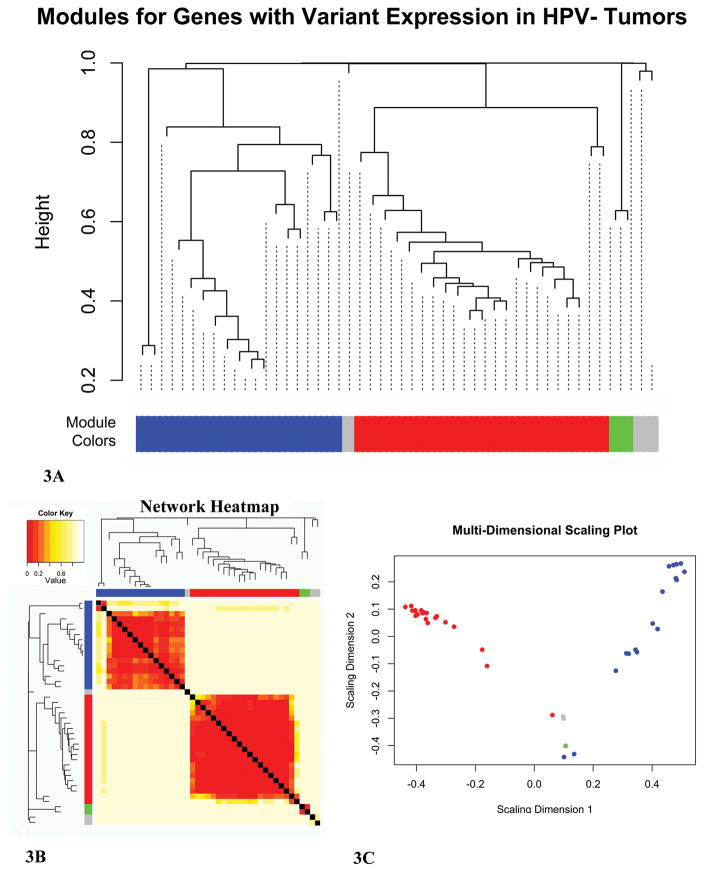
Since the genes with high expression variation in smokers with HPV− tumors potentially represent an important difference between HPV− and HPV+ OPSCC, we analyzed the expression relationships among the selected genes. To gain insight into the possible relationship between the genes with highly variable expression, we used a gene co-expression network approach. This method identifies networks of genes that share expression levels. Our goal was to identify co-expression networks with hub genes that were highly co-expressed with the rest of the network. These hub genes would have the potential to be key players in the breakage of robustness within tumors from HPV− smokers. All 20 HPV− tumors from smokers were used to generate a dendrogram based on the expression values for the 43 selected genes (data not shown). One outlier was identified and removed from all subsequent analyses. A power adjacency matrix was generated for the remaining 19 tumors to establish co-expression relationships among the variably expressed genes. The matrix consists of the coefficient variations for all possible combinations of genes across the patients used in the analysis. The matrix was then raised to a β value to provide weighted relationships which better represents genetic data34. To visualize the co-expression networks, a Topological Overlap Matrix-based dissimilarity (DistTOM) dendrogram was constructed, and the resulting clusters were placed into modules labeled by unique colors (Figure 3A). Two major clusters were identified, shown as blue and red modules. To highlight the gene expression overlap within the identified clusters, a DistTOM heatmap was generated (Figure 3B). A multi-dimensional scaling plot was used as an additional method for visualizing the gene clusters (Figure 3C). Based on a strong DistTOM overlap, as shown by dark coloring on the heatmap and a scattering of genes on the multi-dimensional plot, the blue and red modules formed clear networks of co-expression. The grey module did not form a co-expression network based on a weak DistTOM overlap shown as light coloring on the heatmap and the clustering of genes on the multi-dimensional scaling plot. The green cluster included two transcripts from the same gene and it was not considered for network analysis. The blue and red modules were then used to construct two co-expression networks, which are referred to as the blue and red networks for the remainder of this paper.
Figure 3. Weighted Gene Co-Expression Network (WGCNA) Construction.
Network construction using the WGCNA R package33. A. TOM-based dissimilarity cluster dendrogram of genes with high expression variation in 19 Human Papillomavirus negative (HPV−) tumors from smokers. Gene clusters are assigned to colored modules (blue, grey, red, and green). B. Heatmap representation of the clusters with module colors. Dark red signifies high network overlap between two genes. C. Multi-dimensional scaling visualization of the colored modules. Scattered modules represent strong co-expression networks. Ends of the scattered modules represent hub genes (i.e. the genes with the most connections).
Using the WGCNA package33, we identified two networks (blue and red networks) of co-expression within the 43 selected genes. Once the networks were identified, we assessed the number of connections for each gene within its network to isolate hub genes. The blue co-expression network consisted of 16 variably expressed genes (see Supplemental Table 1 for GO) and included PIGX as a potential hub gene based on a connection weight of 0.4 (Figure 4A). The red co-expression network was slightly larger with 20 genes (see Supplemental Table 2 for GO) and included two potential hub genes, CRIPAK and FOXP4 based on a connection weight of 0.5 (Figure 4B).
Figure 4. Weighted Gene Co-Expression Networks.
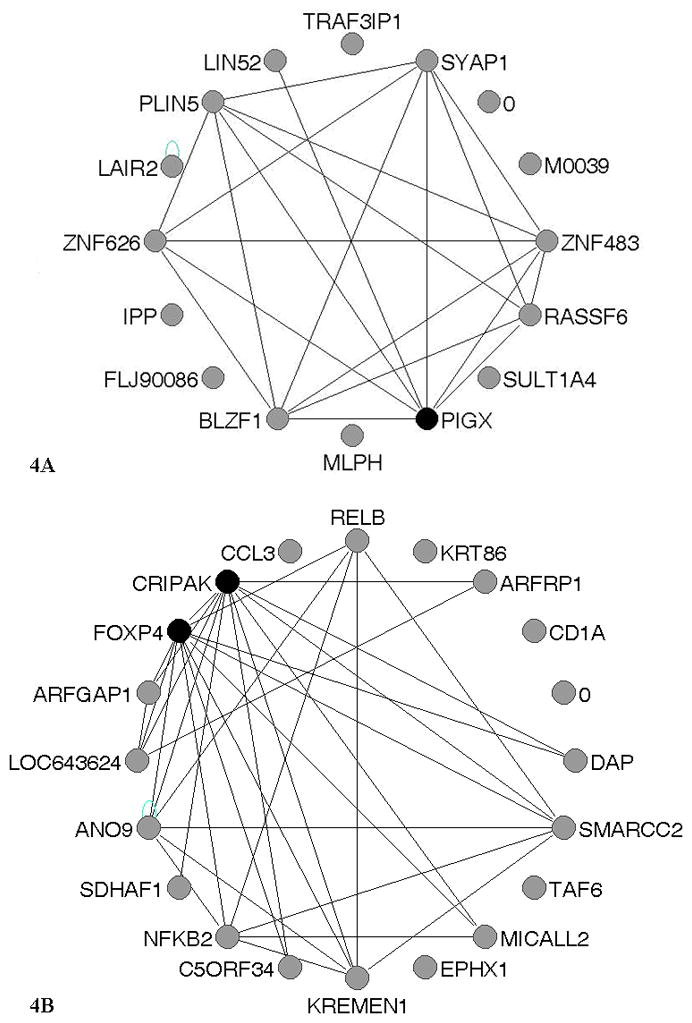
Expression networks were constructed for the blue and red modules. Genes that are colored black are the hub genes (the genes with the most connections within the network). A. The network representation of the blue module. A connection weight cutoff of 0.4 was used within the VisANT software55 to visualize the genes with the most connections and identify the hub gene, PIGX (Phosphatidylinositol Glycan Anchor Biosynthesis, Class X). B. The network representation of the red module. A connection weight cutoff of 0.5 was used to identify two hub genes, CRIPAK (cysteine-rich PAK1 inhibitor) and FOXP4 (forkhead box P4).
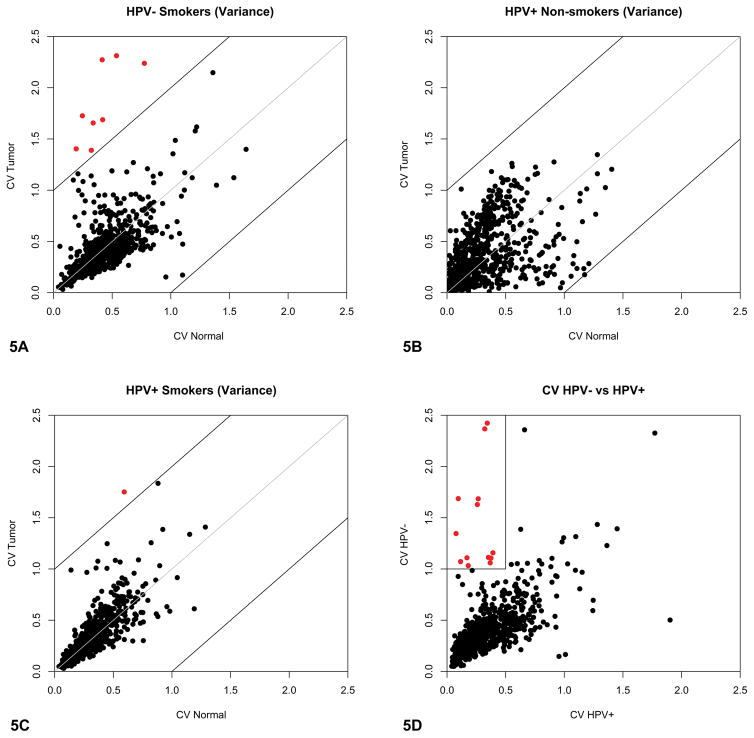
We also assessed whether similar associations with HPV and smoking existed for miRNA expression. Variable levels of typically stable miRNAs in normal tissues could have significant implications during tumorigenesis. As with the mRNA array, we assessed miRNA expression levels in HPV− and HPV+ tumors (see Materials and Methods). Unlike the mRNA microarray, the miRNA array probed far fewer targets. The CVs for all the miRNAs were plotted for tumor and normal tissues, and separated by patient HPV and smoking status. Since there were only two non-smoking patients with HPV− tumors, they were not included in the analyses. The same parameters used for CV analysis in the mRNA dataset were used to analyze the miRNA array data. The miRNAs that had highly variable expression in both tumor and normal tissue samples for each group were highlighted in red. For the smokers with HPV− tumors, there were 8 miRNAs that had high expression variation among the tumors compared to no miRNAs with highly variable expression in the normal tissue samples (Figure 5A). Conversely, there were no miRNAs with highly variable expression in either the tumor or normal tissue for non-smokers with HPV+ tumors (Figure 5B). However, for the smokers with HPV+ tumors, there was one miRNA with high expression variation among the tumors and no miRNAs with highly variable expression in the normal tissue (Figure 5C). Since there were more genes with high expression variation for the smokers, the CV analyses support our first hypothesis that tumors from smokers gain genetic variation during tumorigenesis. Secondly, since there were fewer miRNAs with highly variable expression in the HPV+ tumors of both smokers and non-smokers compared to the HPV− tumors of smokers, the miRNA dataset may support our second hypothesis that virally-driven tumorigenesis is genetically a more controlled process. More HPV− tumors from non-smokers need to be analyzed to fully interpret these results.
Figure 5. Increased microRNA (miRNA) expression in Human Papillomavirus negative (HPV−) tumors from smokers.
A–C. Scatterplots representing individual miRNA coefficients of variation (CV) for tumors and matched normal tissue. Genes with CV>1 both for tumors and for normal tissues are colored red. HPV− tumors from non-smokers are not shown due to a sample size of 2. A. CVs plotted for 14 paired HPV− tumors from smokers. Eight miRNAs had greater CVs in tumor samples compared to no miRNAs in the corresponding normal tissue, suggesting a greater amount of expression variation among tumor miRNAs. B. CVs plotted for 3 paired HPV+ tumors from non-smokers. Conversely, there were no miRNAs with high expression variation in the HPV+ tumors from the non-smoking group. C. CVs plotted for 18 paired HPV+ smokers. Only one miRNA had a high CV in the tumors, while there were no miRNAs with highly variable expression in the normal tissue. D. Scatterplot representing individual miRNA coefficients of variation for 14 HPV− tumors from smokers and 3 HPV+ tumors from non-smokers. Eighteen miRNAs with high expression variation in HPV− tumors from smokers are colored red.
As done with the mRNA expression data, we next compared the HPV− tumors from smokers to the HPV+ tumors from non-smokers (Figure 5D). The CVs for each miRNA were plotted and the miRNAs that had high expression variation among the HPV− tumors were highlighted in red (see Materials and Methods for the parameters used). As expected, there was an observable increase in the number of miRNAs that had highly variable levels of expression and an increase in the range of expression variation for HPV− tumors from smokers compared to the HPV+ tumors from non-smokers. Table 3 lists the 18 miRNAs that had highly variable levels of expression in HPV− tumors along with inferred or experimentally confirmed cellular functions. Unlike mRNAs which are transcribed into a limited number of proteins, miRNAs can target multiple mRNAs in various pathways, resulting in a variety of potential mechanisms by which miRNAs can induce tumorigenesis.
Table 3.
Functions and Interactions for 18 miRNAs with high Expression Variation in HPV− Tumors from Non-smoking Patients.
| miRNA | Cellular Function/Interactions |
|---|---|
| HS_240 | implicated in oesophageal carcinoma |
| hsa-miR-122a | liver specific miRNA |
| hsa-miR-154* | ZNFs are inferred targets |
| hsa-miR-302a | G1/S phase transition |
| hsa-miR-302a* | non-functional |
| hsa-miR-302b | epithelial-mesenchymal transition57 |
| hsa-miR-302d | G1/S phase transition |
| hsa-miR-376b | expressed by endothelial cells exposed to hypoxia58 |
| hsa-miR-431 | binds CD81 (signal transduction) |
| hsa-miR-506 | binds CDK6 (phosphorylates Rb) |
| hsa-miR-507 | colony formation, invasion, growth, and apoptosis (inferred) |
| hsa-miR-508 | MBD1, p53, and Myc are inferred targets |
| hsa-miR-509 | Binds NTRK3 (neuronal differentiation and survival) |
| hsa-miR-510 | binds SPDEF (SAM Pointed Domain Containing ETS Transcription Factor) |
| hsa-miR-513 | association with skin cancer (inferred) |
| hsa-miR-514 | association with skin cancer (inferred) |
| hsa-miR-539 | colony formation, invasion, growth, and apoptosis (inferred) |
| hsa-miR-551b | DNMT3A, TRAF1, and E2F6 are inferred targets |
less prominent form of the miRNA
CD81= Cluster of Differentiation 81
CDK6= Cyclin-Dependent Kinase 6
DNMT3A= DNA (cytosine-5)-Methyltransferase 3A
E2F6= Transcription Factor E2F6
ETS= E26 Transformation-Specific
HPV= Human Papillomavirus
MBD1= Methyl-CpG-Binding Domain Protein 1
miRNA= microRNA
NTRK3= Neurotrophic Tyrosine Kinase, receptor, type 3
p53= tumor protein 53
SAM= Serine Alpha Motif
TRAF1= TNF (Tumor Necrosis Factor) Receptor-Associated Factor 1
ZNF= Zinc Finger proteins
Discussion
We describe a unique model for studying cancer and tumorigenesis. The down-regulation of tumor suppressors and the up-regulation of oncogenes have been the most frequently used method for studying cancer. The identification of tumor suppressors and oncogenes, such as Rb and Ras respectively, lead to the rapid progression of cancer research for tumors where these key regulators induced tumorigenesis39–41. However, many tumors do not exhibit alterations solely in tumor suppressors or oncogenes, but rather they display multiple mutations in the genome. Alternative models of tumorigenesis and tumor progression have been proposed and studied extensively. One approach is focused on the transformation to a more “stem-cell-like” phenotype as a model for cancer progression, which looks for the expression of genes involved in development, motility, and invasion42–44. The stem-like model provides a mechanism for tumor invasion and metastasis, but does not begin to tackle the process of tumorigenesis itself. Additionally, cancer has been modeled through a different form of robustness45, here defined as the capacity of tumor cells to avoid apoptosis46–48. While this approach may be useful for explaining the variance seen among tumors of the same type, it fails to address the mechanism by which normal cells lose robustness of their own complex genotype-phenotype maps. Here we look at cancer as the result of various mechanisms whereby the robustness of normal, non-tumorigenic cells has been broken.
Robustness, which evolved as a result of multiple possible mechanisms, buffers genotypic alterations from manifesting at the phenotypic level16,20,21. That is, robustness allows for the harboring of cryptic genetic variation without disrupting the complex genotypic-phenotypic maps23,24. This enables individuals to exhibit genetic variation without compromising important cellular functions. Under certain stressful conditions, robustness may be lost, exposing the cryptic genetic variation at the phenotypic level. We argue that cancer goes through such a process, whereby increased genetic mutations result in a breakage of robustness leading to tumorigenesis. In our study, we consider gene expression as the most basic phenotype of genetic variation. Thus, we look for an increase in gene expression variation as a result of breakage of robustness. This hypothesis requires the accumulation of a certain number of key mutations to achieve malignant transformation. If a normal cell is primed to become a cancer cell through a different mechanism, such as viral infection by HPV, such loss of robustness may not occur as it does in carcinogen-driven cancers. During virally-induced tumorigenesis, random mutations may be less likely since viral infection of the cell disrupts the cell cycle and apoptotic pathways consistently to promote its own survival. However, we do expect to see some degree of variation among genes from HPV+ tumors since both expression of viral genes and viral integration49 may disrupt a number of important genes thereby leading to a breakage of robustness. There are other models available to study virally-driven tumorigenesis, such as HPV infection in the cervix, Hepatitis B virus infection in the liver, and the role of Epstein-Barr virus infection during B-cell transformation25–27. In this study, we chose OPSCC as a model where both carcinogen- and virally-induced tumorigenesis may occur to test two hypotheses. The first being that tumor cells experience a breakage of robustness, and the second that virally-induced tumorigenesis is a more controlled process. We predicted that tumor samples would exhibit increased gene expression variation with respect to their adjacent, histologically normal tissue. Furthermore, we predicted that smoking related tumors would increase expression variability compared to virally related tumors.
Through our analyses, we identified a number of genes with high expression variation among the HPV− tumors from smokers compared to HPV+ tumors from non-smokers. These genes were divided into networks of co-expression with hub genes, which potentially represent or interact with drivers of phenotypic variance during tumorigenesis. Since we did not include the full microarray for the co-expression network construction, it is likely that the hub genes may not be key drivers. Rather the hubs may represent a downstream player of a key driver of tumorigenesis, or simply share transcription factor regulation with the rest of the network. Epigenetic mechanisms such as altered chromatin marks, DNA methylation, and/or miRNA levels may also be driving network construction. Since these hub genes may not be directly regulating the rest of the network, it is key to look at them within the context of their gene ontology (GO) and pathways to understand their importance in cancer. One network included hub gene PIGX, Phosphatidylinositol Glycan Anchor Biosynthesis, Class X, a key player during Glycosylphosphatidylinositol (GPI-anchor) synthesis50. The GPI-anchor is a post-translational lipid modification added to cell surface transmembrane proteins51. We propose that during tumorigenesis, variation in PIGX expression may perturb the cell signaling and cell-to-cell cross talk of important proteins that are unable to anchor into the plasma membrane. Another network included hub gene CRIPAK, Cysteine-Rich PAK1 Inhibitor, a regulator of cellular senescence through Pak1 inhibition52. We hypothesize that variation in CRIPAK expression may represent a mechanism for tumorigenesis that may occur through dysregulation of senescence. A second hub gene from this network is FOXP4, Forkhead Box P4, a transcription factor important during development53. FOXP4 expression by neural epithelial cells post differentiation suppresses N-cadherin54 suggesting its importance for normal cellular functioning. N-cadherin expression is a marker for epithelial-mesenchymal transition (EMT)55, and we think the variation in FOXP4 expression may represent the breakage of robustness of the epithelial phenotype. For both networks, the GO for the non-hub genes revealed a range of functions for apoptotic, proliferation, cell signaling, and structural genes. These genes warrant further study since their expression values were more variant in HPV− tumors from smokers and could be used as key markers to differentiate between carcinogen- and virally-induced tumorigenesis.
As with gene expression, a number of miRNAs were identified to have high expression variation among the HPV− tumors from smokers compared to HPV+ tumors from non-smokers. We observed very little variation for miRNA expression among HPV+ tumors regardless of smoking status, suggesting that miRNAs are under viral control56. However, we cannot speculate about smoking-induced miRNA expression variation in the absence of viral infection due to the lack of HPV− tumors from non-smoking patients. There are many algorithms to predict potential miRNA targets based on the seed sequences, however the only way to truly identify a real miRNA target or function is through experimental validation57. For this reason, we only discuss two miRNAs with highly variable expression that have been validated in the literature to date. One of the miRNAs with high expression variation, hsa-miR-302b, has been shown to be a regulator of cell reprogramming and targets genes responsible for EMT58. Not only does this miRNA have an important role in tumorigenesis, its identification in our list of variant miRNAs also supports our approach. We propose that variant expression levels of hsa-miR302b indicate a loss of robustness with respect to its targets and complex genotype-phenotype networks. The second miRNA, hsa-miR-376b, has been identified as a miRNA expressed by endothelial cells during hypoxia59, and more recently has been shown to regulate autophagy induced by starvation60. Hypoxia and metabolic starvation are both consequences of tumor growth. The other miRNA functions/interactions have only been inferred and not experimentally validated, however, a few may be relevant to the study of loss of robustness in tumorigenesis and warrant further exploration.
In addition to mRNA and miRNA expression, methylation patterns, copy number variation, and mutation rates for OPSCC should be considered for study using our approach to reveal the underlying mechanisms for gene expression alterations. Recently, a study published by the Cancer Genome Atlas Network (TCGA) reported differences in copy number and somatic mutations between HPV− and HPV+ HNSCCs61. TCGA findings also revealed differential methylation patterns, copy number variation, and somatic mutations between HPV+ HNSCCs with integrated and non-integrated genomes, which may indicate varying mechanisms of HPV-driven tumorigenesis61,62. Finally, although we identified genes and pathways that have not been previously associated with a loss of robustness in cancer cells, these findings need to be corroborated experimentally and additional analyses of what might have contributed to increased gene expression variation need to be done. As we have seen in yeast knockouts, not all disruptions result in increased phenotypic variation21. What would be required for a better understanding would be to identify the characteristics of genes that increase phenotypic variation when either mutated or disrupted epigenetically. Characteristics in this case, are defined as the biological functions, downstream targets, and position in the network hierarchy that are relevant for the progression of cancer. Once identified, these characteristics may reveal novel underlying driving mechanisms for the cancer phenotype.
Supplementary Material
Acknowledgments
Support
This work is supported in part by National Institute of Dental & Craniofacial Research and National Cancer Institute grants (CA131648, CA115243 and DE023941 to N. F. S. and T. J. B.) and 1-R01-AG028872-01A1 (to AB).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: a cancer journal for clinicians. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Jaber MA, Porter SR, Gilthorpe MS, Bedi R, Scully C. Risk factors for oral epithelial dysplasia--the role of smoking and alcohol. Oral oncology. 1999;35(2):151–156. doi: 10.1016/s1368-8375(98)00106-7. [DOI] [PubMed] [Google Scholar]
- 3.Pytynia KB, Dahlstrom KR, Sturgis EM. Epidemiology of HPV-associated oropharyngeal cancer. Oral oncology. 2014;50(5):380–386. doi: 10.1016/j.oraloncology.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreimer AR, Clifford GM, Boyle P, Franceschi S. Human papillomavirus types in head and neck squamous cell carcinomas worldwide: a systematic review. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2005;14(2):467–475. doi: 10.1158/1055-9965.EPI-04-0551. [DOI] [PubMed] [Google Scholar]
- 5.Vidal L, Gillison ML. Human papillomavirus in HNSCC: recognition of a distinct disease type. Hematology/oncology clinics of North America. 2008;22(6):1125–1142. vii. doi: 10.1016/j.hoc.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Schlecht NF, Burk RD, Adrien L, et al. Gene expression profiles in HPV-infected head and neck cancer. The Journal of pathology. 2007;213(3):283–293. doi: 10.1002/path.2227. [DOI] [PubMed] [Google Scholar]
- 7.Troy JD, Weissfeld JL, Youk AO, Thomas S, Wang L, Grandis JR. Expression of EGFR, VEGF, and NOTCH1 suggest differences in tumor angiogenesis in HPV-positive and HPV-negative head and neck squamous cell carcinoma. Head and neck pathology. 2013;7(4):344–355. doi: 10.1007/s12105-013-0447-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Kempen PM, Noorlag R, Braunius WW, Stegeman I, Willems SM, Grolman W. Differences in methylation profiles between HPV-positive and HPV-negative oropharynx squamous cell carcinoma: A systematic review. Epigenetics. 2014;9(2):194–203. doi: 10.4161/epi.26881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lleras RA, Smith RV, Adrien LR, et al. Unique DNA methylation loci distinguish anatomic site and HPV status in head and neck squamous cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(19):5444–5455. doi: 10.1158/1078-0432.CCR-12-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lajer CB, Garnaes E, Friis-Hansen L, et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. British journal of cancer. 2012;106(9):1526–1534. doi: 10.1038/bjc.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bol V, Gregoire V. Biological basis for increased sensitivity to radiation therapy in HPV-positive head and neck cancers. BioMed research international. 2014;2014:696028. doi: 10.1155/2014/696028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirghani H, Amen F, Blanchard P, et al. Treatment de-escalation in HPV-positive oropharyngeal carcinoma: Ongoing trials, critical issues and perspectives. International journal of cancer. Journal international du cancer. 2015;136(7):1494–1503. doi: 10.1002/ijc.28847. [DOI] [PubMed] [Google Scholar]
- 13.Salazar CR, Anayannis N, Smith RV, et al. Combined P16 and human papillomavirus testing predicts head and neck cancer survival. International journal of cancer. Journal international du cancer. 2014;135(10):2404–2412. doi: 10.1002/ijc.28876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nygard M, Aagnes B, Bray F, Moller B, Mork J. Population-based evidence of increased survival in human papillomavirus-related head and neck cancer. European journal of cancer. 2012;48(9):1341–1346. doi: 10.1016/j.ejca.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 15.Sethi S, Ali-Fehmi R, Franceschi S, et al. Characteristics and survival of head and neck cancer by HPV status: a cancer registry-based study. International journal of cancer. Journal international du cancer. 2012;131(5):1179–1186. doi: 10.1002/ijc.26500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masel J, Siegal ML. Robustness: mechanisms and consequences. Trends in genetics : TIG. 2009;25(9):395–403. doi: 10.1016/j.tig.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waddington CH. Canalization of Development and the Inheritance of Acquired Characters. Nature. 1942;(150):563–565. [Google Scholar]
- 18.Waddington CH. The Strategy of the Genes. London: Allen & Unwin; 1957. [Google Scholar]
- 19.Waddington CH. Evolutionary Systems–Animal and Human. Nature. 1959;(183):1634–1638. doi: 10.1038/1831634a0. [DOI] [PubMed] [Google Scholar]
- 20.Siegal ML, Bergman A. Waddington’s canalization revisited: developmental stability and evolution. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(16):10528–10532. doi: 10.1073/pnas.102303999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergman A, Siegal ML. Evolutionary capacitance as a general feature of complex gene networks. Nature. 2003;424(6948):549–552. doi: 10.1038/nature01765. [DOI] [PubMed] [Google Scholar]
- 22.Belbin TJ, Bergman A, Brandwein-Gensler M, et al. Head and neck cancer: reduce and integrate for optimal outcome. Cytogenetic and genome research. 2007;118(2–4):92–109. doi: 10.1159/000108290. [DOI] [PubMed] [Google Scholar]
- 23.Paaby AB, Rockman MV. Cryptic genetic variation: evolution’s hidden substrate. Nature reviews. Genetics. 2014;15(4):247–258. doi: 10.1038/nrg3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson G, Dworkin I. Uncovering cryptic genetic variation. Nature reviews. Genetics. 2004;5(9):681–690. doi: 10.1038/nrg1426. [DOI] [PubMed] [Google Scholar]
- 25.Hawley-Nelson P, Vousden KH, Hubbert NL, Lowy DR, Schiller JT. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. The EMBO journal. 1989;8(12):3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paterlini-Brechot P, Saigo K, Murakami Y, et al. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene. 2003;22(25):3911–3916. doi: 10.1038/sj.onc.1206492. [DOI] [PubMed] [Google Scholar]
- 27.Price AM, Luftig MA. Dynamic Epstein-Barr virus gene expression on the path to B-cell transformation. Advances in virus research. 2014;88:279–313. doi: 10.1016/B978-0-12-800098-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Williams V, Filippova M, Filippov V, Duerksen-Hughes P. Viral carcinogenesis: factors inducing DNA damage and virus integration. Cancers. 2014;6(4):2155–2186. doi: 10.3390/cancers6042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Freitas LB, Burk RD. Evolution and classification of oncogenic human papillomavirus types and variants associated with cervical cancer. Methods in molecular biology. 2015;1249:3–26. doi: 10.1007/978-1-4939-2013-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leek JTJW, Parker HS, Fertig EJ, Jaffe AE, Storey JD. R package version 3.12.0. sva: Surrogate Variable Analysis. [Google Scholar]
- 31.Sultan M, Piccini I, Balzereit D, et al. Gene expression variation in Down’s syndrome mice allows prioritization of candidate genes. Genome biology. 2007;8(5):R91. doi: 10.1186/gb-2007-8-5-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang HGJ. supraHex: an R/Bioconductor package for tabular omics data analysis using a supra-hexagonal map. Biochemical and biophysical research communications. 2014;443(1):285–289. doi: 10.1016/j.bbrc.2013.11.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Statistical applications in genetics and molecular biology. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 35.Hu Z, Mellor J, Wu J, DeLisi C. VisANT: an online visualization and analysis tool for biological interaction data. BMC bioinformatics. 2004;5:17. doi: 10.1186/1471-2105-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome research. 2003;13(9):2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris T, Jimenez L, Kawachi N, et al. Low-level expression of miR-375 correlates with poor outcome and metastasis while altering the invasive properties of head and neck squamous cell carcinomas. The American journal of pathology. 2012;180(3):917–928. doi: 10.1016/j.ajpath.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 39.Murphree AL, Benedict WF. Retinoblastoma: clues to human oncogenesis. Science. 1984;223(4640):1028–1033. doi: 10.1126/science.6320372. [DOI] [PubMed] [Google Scholar]
- 40.Parada LF, Tabin CJ, Shih C, Weinberg RA. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297(5866):474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- 41.Weinberg RA. Oncogenes and tumor suppressor genes. CA: a cancer journal for clinicians. 1994;44(3):160–170. doi: 10.3322/canjclin.44.3.160. [DOI] [PubMed] [Google Scholar]
- 42.Ksiazkiewicz M, Markiewicz A, Zaczek AJ. Epithelial-mesenchymal transition: a hallmark in metastasis formation linking circulating tumor cells and cancer stem cells. Pathobiology : journal of immunopathology, molecular and cellular biology. 2012;79(4):195–208. doi: 10.1159/000337106. [DOI] [PubMed] [Google Scholar]
- 43.Chen C, Zimmermann M, Tinhofer I, Kaufmann AM, Albers AE. Epithelial-to-mesenchymal transition and cancer stem(-like) cells in head and neck squamous cell carcinoma. Cancer letters. 2013;338(1):47–56. doi: 10.1016/j.canlet.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 44.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nature reviews. Molecular cell biology. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaneko K. Characterization of stem cells and cancer cells on the basis of gene expression profile stability, plasticity, and robustness: dynamical systems theory of gene expressions under cell-cell interaction explains mutational robustness of differentiated cells and suggests how cancer cells emerge. BioEssays : news and reviews in molecular, cellular and developmental biology. 2011;33(6):403–413. doi: 10.1002/bies.201000153. [DOI] [PubMed] [Google Scholar]
- 46.Tian T, Olson S, Whitacre JM, Harding A. The origins of cancer robustness and evolvability. Integrative biology : quantitative biosciences from nano to macro. 2011;3(1):17–30. doi: 10.1039/c0ib00046a. [DOI] [PubMed] [Google Scholar]
- 47.Masuda M, Toh S, Wakasaki T, Suzui M, Joe AK. Somatic evolution of head and neck cancer - biological robustness and latent vulnerability. Molecular oncology. 2013;7(1):14–28. doi: 10.1016/j.molonc.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radisavljevic Z. AKT as locus of cancer positive feedback loops and extreme robustness. Journal of cellular physiology. 2013;228(3):522–524. doi: 10.1002/jcp.24167. [DOI] [PubMed] [Google Scholar]
- 49.Akagi K, Li J, Broutian TR, et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome research. 2014;24(2):185–199. doi: 10.1101/gr.164806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashida H, Hong Y, Murakami Y, et al. Mammalian PIG-X and yeast Pbn1p are the essential components of glycosylphosphatidylinositol-mannosyltransferase I. Molecular biology of the cell. 2005;16(3):1439–1448. doi: 10.1091/mbc.E04-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chatterjee S, Mayor S. The GPI-anchor and protein sorting. Cellular and molecular life sciences : CMLS. 2001;58(14):1969–1987. doi: 10.1007/PL00000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Talukder AH, Meng Q, Kumar R. CRIPak, a novel endogenous Pak1 inhibitor. Oncogene. 2006;25(9):1311–1319. doi: 10.1038/sj.onc.1209172. [DOI] [PubMed] [Google Scholar]
- 53.Teufel A, Wong EA, Mukhopadhyay M, Malik N, Westphal H. FoxP4, a novel forkhead transcription factor. Biochimica et biophysica acta. 2003;1627(2–3):147–152. doi: 10.1016/s0167-4781(03)00074-5. [DOI] [PubMed] [Google Scholar]
- 54.Rousso DL, Pearson CA, Gaber ZB, et al. Foxp-mediated suppression of N-cadherin regulates neuroepithelial character and progenitor maintenance in the CNS. Neuron. 2012;74(2):314–330. doi: 10.1016/j.neuron.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hazan RB, Phillips GR, Qiao RF, Norton L, Aaronson SA. Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. The Journal of cell biology. 2000;148(4):779–790. doi: 10.1083/jcb.148.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salazar C, Calvopina D, Punyadeera C. miRNAs in human papilloma virus associated oral and oropharyngeal squamous cell carcinomas. Expert review of molecular diagnostics. 2014;14(8):1033–1040. doi: 10.1586/14737159.2014.960519. [DOI] [PubMed] [Google Scholar]
- 57.Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44(1):47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Subramanyam D, Lamouille S, Judson RL, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nature biotechnology. 2011;29(5):443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voellenkle C, Rooij J, Guffanti A, et al. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. Rna. 2012;18(3):472–484. doi: 10.1261/rna.027615.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Korkmaz G, le Sage C, Tekirdag KA, Agami R, Gozuacik D. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy. 2012;8(2):165–176. doi: 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- 61.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parfenov M, Pedamallu CS, Gehlenborg N, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(43):15544–15549. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.