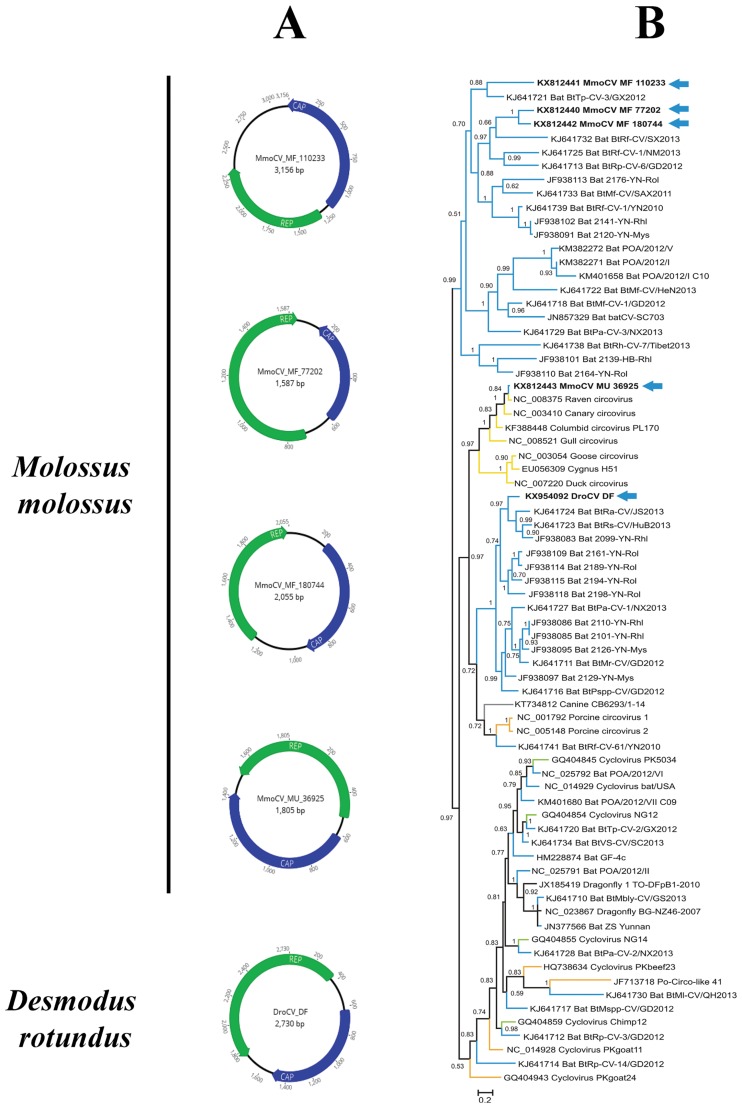
Fig 3. Circular genome maps of five putative circoviruses directly recovered from metagenomic data of M. molossus and D. rotundus bat feces and their phylogenetic relationships with other representative members of the Circoviridae family.
(A) The inversely arranged open-reading frames encoding the putative replication-associated protein (REP) and capsid protein (CAP) are shown in green and blue boxes, respectively. The genome organization was determined with Geneious R9. (B) The phylogenetic analysis is based on the REP protein sequences (alignment of 109 amino acids). The blue arrows indicate the five REP sequences of bat-sourced circoviruses obtained in the present study. The tree was inferred using the Bayesian method with the blosum62 model. Sequence identifiers include the NCBI accession number and the isolate name. Posterior probabilities of the Bayesian analysis (>50%) are shown next to each node. The scale bar indicates amino acid substitutions per site.