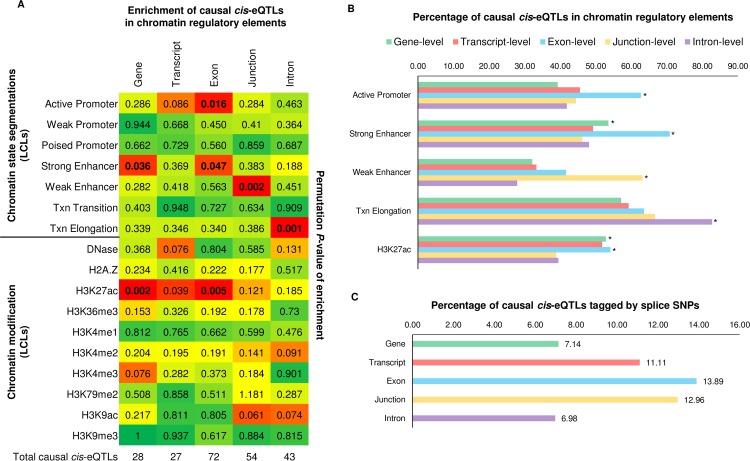
Fig 6. Functional annotation of causal autoimmune cis-eQTLs.
(A) We took the causal autoimmune cis-eQTLs detected for each RNA-Seq quantification type and performed enrichment testing for chromatin state segmentation and histone marks in LCLs taken from the NIH Roadmap Epigenomics Project. We used the GoShifter algorithm to do this (see methods); which takes all SNPs in strong LD (r2>0.8) with the causal cis-eQTLs and calculates the proportion of SNPs overlapping chromatin marks, the positions of the marks are then shuffled whilst retaining the SNP positions, and the fraction of overlap recalculated over 1,000 permutations. A permutation P-value is then generated–which is annotated in each box (P<0.05 deemed significant). The heat colour is representative of the permutation P-value. Significant enrichment tests are highlighted in bold. The total number of causal cis-eQTLs per quantification type are annotated at the bottom of the heatmap. (B) The percentage of causal cis-eQTLs in chromatin regulatory marks per quantification type. An asterisk shows that this level of enrichment is deemed to be significant as shown in panel A. (C) The percentage of causal cis-eQTLs in chromatin regulatory marks per quantification type that are or are highly correlated (r2>0.8) with SNPs that alter splice site consensus sequences of the target genes (assessed by Sequence Ontology for the hg19 GENCODE v12 reference annotation).