Figure 8. Different models for histone inheritance.
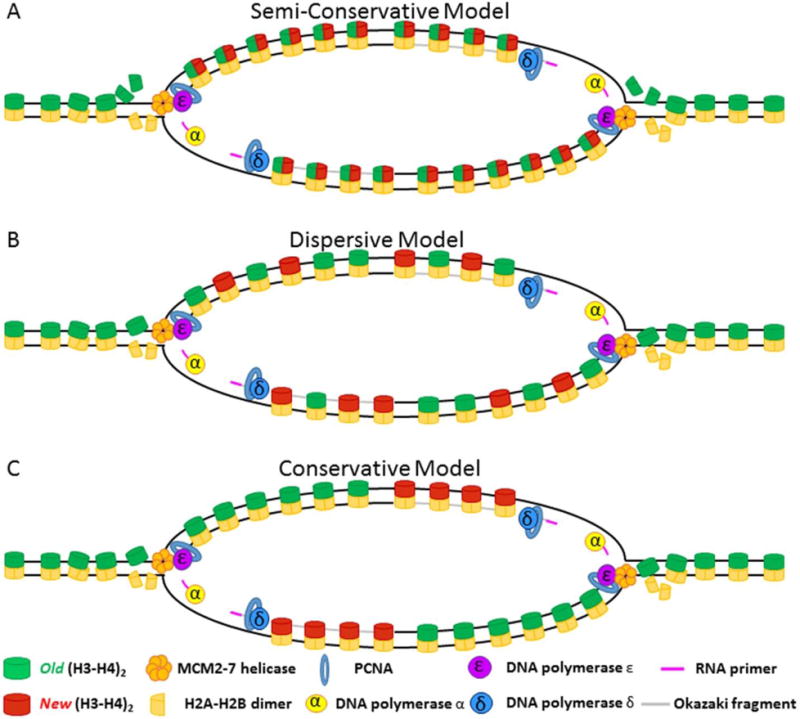
(A) Semi-conservative model: Old (H3–H4)2 tetramers are split at the replication fork, allowing (H3–H4) dimers to be inherited equally on both the leading and the lagging strand. New (H3–H4) dimers pair with old (H3–H4) dimers to recreate the tetramer structure. (B) Dispersive model: Old (H3–H4)2 tetramers remain together at the fork and are randomly segregated to leading or lagging strand in roughly equal numbers. New histones fill in gaps left by old histones to reconstitute nucleosome density. (C) Conservation model: Old (H3–H4)2 tetramers remain un-split at the fork and are inherited as a tetramer. In this model, tetramers are biased in their inheritance such that either the leading strand (shown) or the lagging strand (not shown) inherits a majority of the old (H3–H4)2 tetramers. New histones fill in gaps left by old histones to reconstitute nucleosome density.
