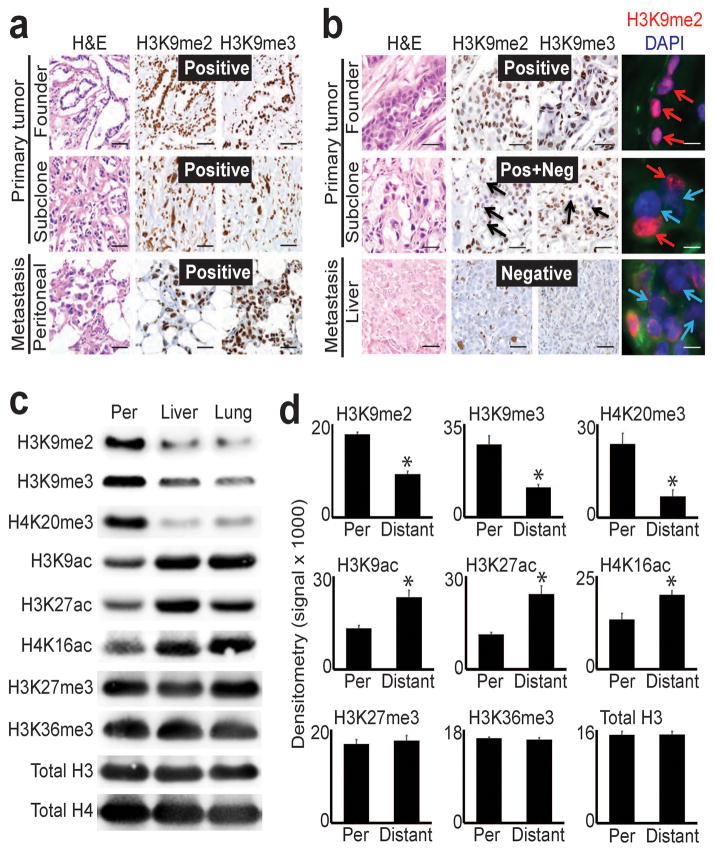
Figure 1. Global epigenetic reprogramming during the evolution of distant metastasis.
(a) IHC stains against H3K9me2/3 performed on tumor sections as indicated for patient A124 (regional/peritoneal spread), which corresponds to samples detailed in Supplementary Table 1 (founder: A124PrF; subclone: A124PrS; peritoneal metastasis: A124Per). Staining was diffusely positive (brown nuclei) across all tumor sections. (b) IHC and IF stains on tumor sections from patient A125 (distant metastatic spread), which corresponds to samples detailed in Supplementary Table 1 (founder: A125PrF; subclone: A125PrS; liver metastasis: A125Lv1). There was progressive loss of nuclear staining that initiated in primary tumor subclones that seeded metastases (middle panels) with diffuse loss in the liver metastases (scale bars=100μm for IHC, 20μm for IF; IF colors: red=H3K9me2; blue=DAPI; green=vimentin). (c) Western blots on cells lines collected from a peritoneal subclone (Per) and distant (liver, lung) metastatic subclones from same patient (pA38, see Supplementary Fig. 1c for corresponding IHC) also showed reduced levels of heterochromatin modifications with increased acetylation in distant metastatic subclones compared to peritoneal. (d) Densitometry summary of western blot findings for the indicated histone modifications across cell lines from distant metastatic subclones compared to peritoneal carcinomatosis (derived from blots shown in Supplementary Fig. 1e–f, n=8 biological replicates, error bars: s.e.m., *p<0.01 by two-tailed t-tests).