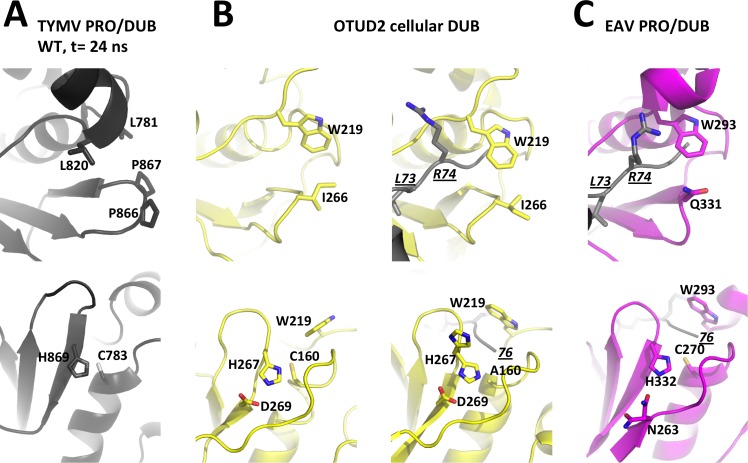
Fig 8. Layouts of the two sides of the active site clefts in viral OTU PRO/DUBs and cellular OTU DUBs.
For each enzyme, the top panel is the view from the P side with residues bridging the hydrophobic constriction as sticks and labeled. The bottom panel is the view from the P' side with residues known to contribute to catalysis as sticks and labeled. For these DUBs for which structures are available in both free and Ub-bound forms (see also S5 and S6 Figs), the free enzyme is displayed on the left, and the complex on the right. In complexes, Ub is in grey with its C-terminal residues 73-LRGG-76 as sticks. L73 and R74 are labeled in underlined italics in one view, and the C-terminus is labeled with an italicized and underlined '76' in the other view. (A) TYMV PRO/DUB. A snapshot at t = 24 ns from the molecular dynamics simulation of the wild type molecule was used here. This snapshot represents the typical equilibrium conformation where the GPP flap is in the intermediate position (Fig 2A, top and Fig 8A, top) and the catalytic dyad realigned (Fig 2A, bottom and Fig 8A, bottom). For clarity, hydrogens added for the simulation are omitted. (B) The cellular OTU DUB domain OTUD2 in free (PDB 4BOQ) and Ub-bound (PDB 4BOS) forms. The latter is a catalytic cysteine mutant C160A and its catalytic histidine displays static disorder with alternate canonical and swung out conformations. (C) The arterivirus PRO/DUB in complex with Ub (PDB 4IUM). Despite its divergence from other OTU DUBs, this enzyme displays a canonical OTU DUB active site cleft both on the P and P' sides. The only exception is the third catalytic residue (here N263) that comes from a neighbouring strand.