Abstract
In this issue of Developmental Cell, Fong et al. (2014) presents evidence for a model of centriole duplication where by the cartwheel – the starting building block in centriole biogenesis – assembles with in the lumen of the mother centriole before templating the daughter centriole to ensure a single duplication event per cell cycle.
Residing at the core of the centrosome, the major microtubule-organizing center (MTOC) in animal cells, is a pair of centrioles. An aesthetic marvel of nature’s engineering, the centriole is a cylinder with stable microtubule triplets arranged in a 9-fold radially symmetric pattern (Figure 1). This precise geometry of the centriole structure and the exact number of centrioles per cell, resulting from a single duplication event per cell cycle, are critical for normal cellular function and development. Indeed, abnormalities in centriole structure, function, and number are associated with ciliopathies, primary microcephalies, primordial dwarfisms, and cancer (Nigg and Raff, 2009),.
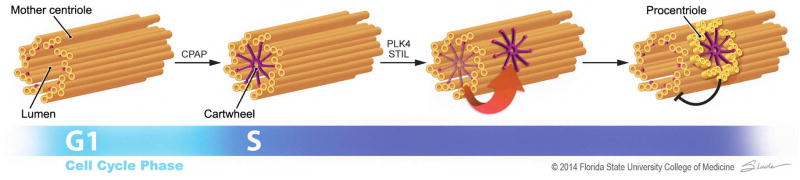
Figure 1. New model for centriole duplication.
In this model, the cartwheel (purple) is preassembled in the lumen of the mother centriole (gold)in a CPAP/SAS-4-dependent manner during S phase. This scaffold is then transferred to the outside surface of the mother through PLK4 and STIL/SAS-5 to initiate daughter centriole assembly. The nascent, engaged daughter centriole blocks additional cartwheel preassembly in the mother, preventing further duplication. Illustration by Jodi Slade.
Centriole biogenesis occurs by at least two pathways. The primary, canonical pathway is where a daughter centriole assembles at the proximal surface of the pre-existing “mother” centriole, suggesting a template-based process. The other pathway is ade novo assembly pathway, which occurs without a pre-existing centriole(Brito et al., 2012). The former assembly mechanism prevails individing cells, where centriole duplication occurs once and only once for each maternal centriole per cell cycle.
The initiation of centriole duplication typically starts with the formation of a 9-fold symmetric cartwheel structure on the side of the preexisting centriole at its proximal base. A nascent centriole then grows on this cartwheel platform (Azimzadeh and Marshall, 2010). The main structural component of the cartwheel is a coiled-coil protein called SAS-6 that dimerizes and then self-assembles into a nine-fold radial cartwheel (Schuldt, 2011). In many cell types, during maturation of the new centriole, the cartwheel structure degenerates, leaving the centriole lumen empty (Azimzadeh and Marshall, 2010). For every rule, however, there are exceptions. For example, in butterfly spermatocytes, the mother centriole retains the cartwheel structure after S phase, and multiple procentrioles(nascent centrioles) are seen next to the assembled daughter, although their assembly is arrested (Gottardo et al., 2014).
Centrioles are licensed to duplicate only once during the typical cell cycle through the disengagement of the mother-daughter centriole pair at the end of mitosis (Tsou and Stearns, 2006). A notable exception to this is the amplification of centrioles that occurs in multiciliated epithelial cells, which produce hundreds of centrioles to template the assembly of a multitude of motile cilia. The overexpression of the major cartwheel component SAS-6, its partner STIL/SAS-5, or the polo-like kinase 4 (PLK4), also cause centrioles to overduplicate. When PLK4 is overexpressed, instead of one daughter forming at the proximal base of a mother centriole, five or six can form around the base of the mother (Kleylein-Sohn et al., 2007). Centriole amplification is a hallmark of cancer cells and can induce tumorigenesis (Nigg and Raff, 2009). In normal dividing cells, however, centriole replication occurs with high fidelity. How is this controlled?
In this issue of Developmental Cell, Fong et al. (2014) propose a new model for centriole replication where by the cartwheel assembles in the lumen of the mother centriole, and then transits to the proximal outer wall to initiate daughter procentriole assembly.
Taking advantage of super-resolution microscopy, Fong et al. (2014) uncovered detailed centriolar localization changes for SAS-6 through the duplication cycle. While confirming previous findings that SAS-6 is destroyed in G1 phase and forms foci in duplicated centrioles (Strnad et al., 2007), the authors observed that a small amount of SAS-6 is transiently recruited to the proximal lumen of unduplicated centrioles in early S phase where it appears to form a cartwheel prior to duplication. This occurs even with a SAS-6 mutant that is unable to self-oligomerize, indicating that the lumen of the mother centriole fosters cartwheel assembly independent of SAS-6’s inherent ability to self-assemble. Moreover, CPAP/SAS-4, previously shown to be required in the early stage of centriole duplication, is required for SAS-6 recruitment to the lumen of the mother centriole. A role for another early assembly factor, CEP135/Bld10, however, was unresolved as its depletion by RNAi was incomplete.
What is the fate of the luminal SAS-6? Fong et al.(2014) demonstrated that after acquiring the apparent cartwheel at the proximal lumen of the mother centriole, the SAS-6 oligomer is released. Concurrent with its release, a new signal for SAS-6 appears on the outside wall at the proximal base of the mother centriole, where a procentriole assembles. While not demonstrated directly, these observations suggest that the cartwheel assembles in the “empty” lumen of the mother centriole in early S phase and once released, transits to the centriole outer wall to initiate procentriole formation (Figure 1). Fong et al. (2014) show that PLK4 and STIL/SAS-5 are both required for the release of SAS-6 from the mother centriole and the appearance of SAS-6 on the outer wall. They also reveal that centriole engagement regulates the recruitment of SAS-6 to the centriole lumen. Thus, when a daughter procentriole assembles, further SAS-6 recruitment to the lumen of the mother centriole is blocked. This regulation ensures that cartwheels preassemble indisengaged centrioles in early Sphase to promote efficient centriole duplication, while also prohibiting any reduplication of engaged centrioles before mitosis.
Together, the work by Fong et al. (2014) provides an attractive new model to explain how centrioles duplicate only once during each centriole replication cycle. By using the mother’s lumen as a “womb” to incubate cartwheel assembly and then transplanting it to her outside surface, this can conceptually explain how only one daughter procentrioleis templated by a mother centriole, although the mechanism by which the procentriole blocks the formation of a new precursor in the lumen of the mother remains unknown. It is notable that during de novo centriole assembly, when there is no mother to template cartwheel assembly, the number of centrioles assembled is highly variable (Loncarek and Khodjakov, 2009). The next challenge for centrosome scientists is to directly show that the cartwheel formed in the mother centriole lumen is the same one that later assembles the daughter centriole on the side of the mother centriole. Furthermore, while the essential role for PLK4 might be cartwheel release from the mother’s lumen as revealed by Fong et al. (2014), it is also required for de novo centriole assembly (Nigg and Raff, 2009), so PLK4 must have additional functions in this context. Nevertheless, the provocative new model that emerges from the work by Fong et al. can be tested further, and may finally begin to explain the fidelity of centriole duplication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copy editing, type setting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azimzadeh J, Marshall WF. Curr Biol. 2010;20(18):R816–25. doi: 10.1016/j.cub.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Gouveia SM, Bettencourt-Dias M. Curr Opin Cell Biol. 2012;24(1):4–13. doi: 10.1016/j.ceb.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Fong CS, Kim M, Yang TT, Liao JC, Tsou MFB. Dev Cell. 2014:30. doi: 10.1016/j.devcel.2014.05.008. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardo M, Callaini G, Riparbelli MG. J Cell Sci. 2014:127. doi: 10.1242/jcs.152843. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Dev Cell. 2007;13(2):190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Loncarek J, Khodjakov A. Mol Cells. 2009;27(2):135–42. doi: 10.1007/s10059-009-0017-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Cell. 2009;139(4):663–78. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Schuldt A. Nat Rev Mol Cell Biol. 2011;12(3):137. doi: 10.1038/nrm3071. [DOI] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Dev Cell. 2007;13(2):203–13. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Nature. 2006;442(7105):947–51. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]