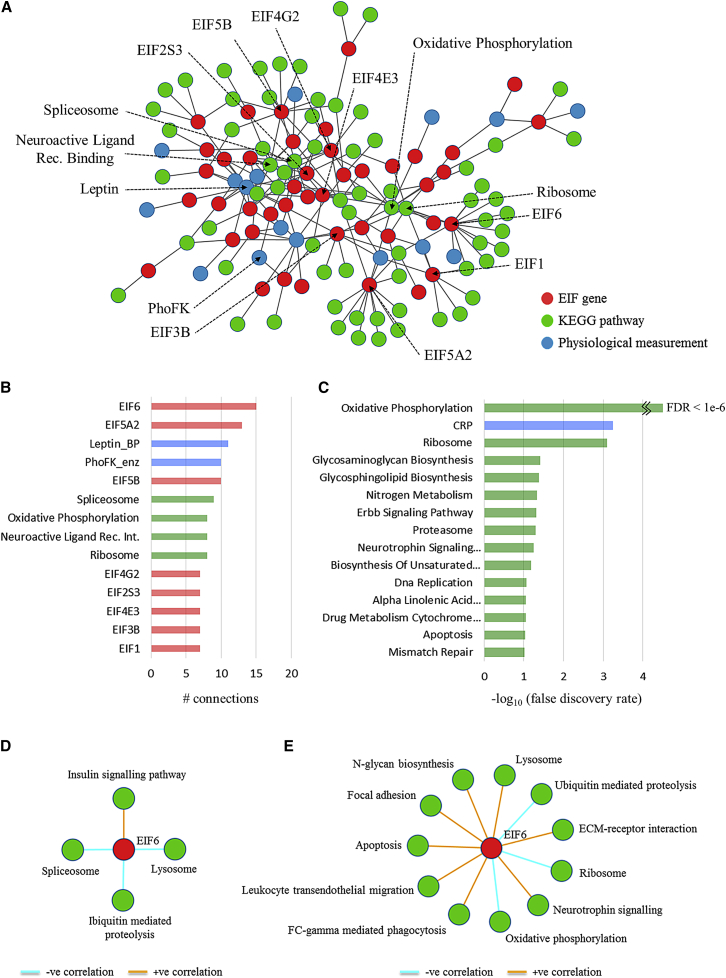
Figure 2.
Network-Based Integration of EIF Genes, Physiological Measurements, and Pathway Activity Reveals Eif6 as the Most Connected Factor
(A) This network represents the integration of multiple analyses. Connections between eIF genes and physiological measurements represent regression analysis using baseline eIF gene expression as a predictor variable (FDR < 10%). Connections between eIF genes and KEGG pathways represent significant enrichment of genes from a KEGG pathway within genes highly differentially co-expressed with the eIF.
(B) The total number of connections (degree) of the top 14 most highly connected components of the network (degree > 6).
(C) Significance of enrichment of KEGG pathways and phenotypic measurements connected to Eif6 in the network.
(D and E) Network visualization of KEGG pathways enriched (FDR < 10%) within genes significantly correlated (FDR < 5%) with Eif6 expression in untrained (n = 2,177 genes) (D) and trained (n = 2,022 genes) (E) individuals separately.