Figure 2. Modification of Lipid A by tailoring enzymes.
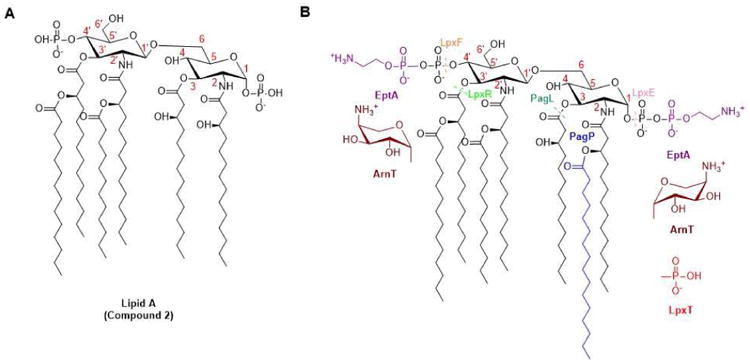
(Adapted from [4]) (A) The canonical structure of Lipid A from E. coli is shown absent the two Kdo sugars, which are not essential for the immunological activities of Lipid A. (B) Modification(s) introduced by each representative tailoring enzyme is indicated in a different color, while components of E. coli Lipid A remain in black. Tailoring enzymes such as ArnT, EptA, LpxT, and PagP catalyze group transfer reactions, while enzymes such as PagL, LpxE, LpxF, and LpxR promote hydrolysis (dashed line across scissile bond). LpxE and/or LpxF homologs are observed in Francisella tularensis[67], Helicobacter pylori [68], Porphyromonas gingivalis [69], Capnocytophaga canimorsus [7], and Bacteroides thetaiotaomicron [70]. LpxR homologs are found in Salmonella typhimurium [71], Yersinia enterocolitica [72], and Helicobacter pylori [73]. PagL is found in Salmonella typhimurium [74] and Pseudomonas aeruginosa [75]. LpxT is found in Escherichia coli [76]. EptA in Escherichia coli [77] and Campylobacter jejuni [78] has broad specificity for both 1 and 4′ positions, while P. aeruginosa EptA only modifies the 4′-phosphate group [79]. PagP in Pseudomonas aeruginosa transfers a palmitoyl chain to the 3-OH of the 3′- acyl residue [80], whereas its homologs in Escherichia coli [81] and Salmonella enterica recognize the 3-OH of the 2-position acyl chain [82,83], and Bordetella parapertussis PagP can modify either hydroxyl group [84]. ArnT in Salmonella typhimurium can decorate the 1- and 4′-phosphates of Lipid A with 4-amino-4-deoxy-L-arabinose (L-Ara4N) [85,86], whereas ArnT in Francisella tularensis adds a galactosamine unit onto the 1-phosphate [87] and ArnT in Bordetella pertussis can add a glucosamine unit on either phosphate groups [88].
