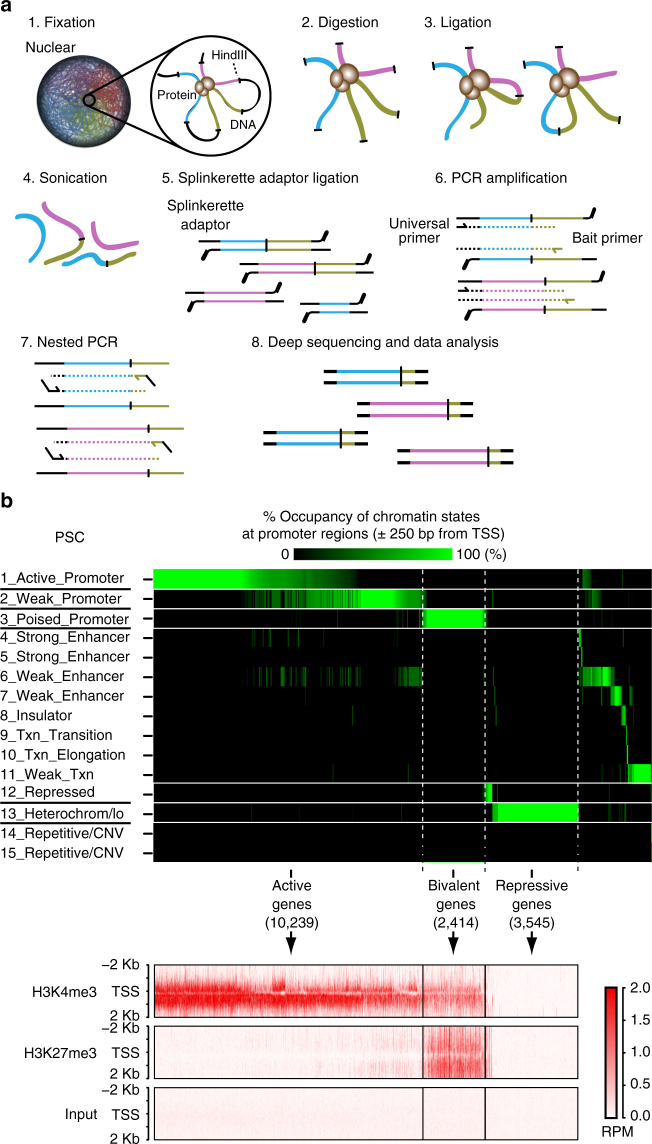
Fig. 1.
ms4C-seq and definition of genes by chromatin states. a Schematic diagram of ms4C-seq. Step 1: fixation of a nucleus with formaldehyde. Step 2: digestion of DNA with HindIII restriction enzyme. Step 3: preparation of chimeric fragments that are derived from physical proximity ligation of DNA. Step 4: fragmentation of those chimeric fragments. Step 5–7: ligations of Splinkerette adapter22 and amplifications of target loci by nested PCR with a universal primer for the adaptor and specific primers designed for bait loci. Step 8: deep sequencing by a next-generation sequencer. See also Supplementary Table 1 and Methods for details. b Definition of transcriptional states of 18,984 genes (bivalent, active, repressive, and other gene groups). The top heat map represents the occupancy of each chromatin state (%) within TSS ± 250 bp for all refseq genes in PSCs (ESCs). Bottom heatmaps indicate H3K4me3 and H3K27me3 modification profiles (RPM values) around TSS for active, repressive, and bivalent genes defined by chromatin states in PSCs (ESCs). Histone modification profiles are shown within TSS ± 2 Kb