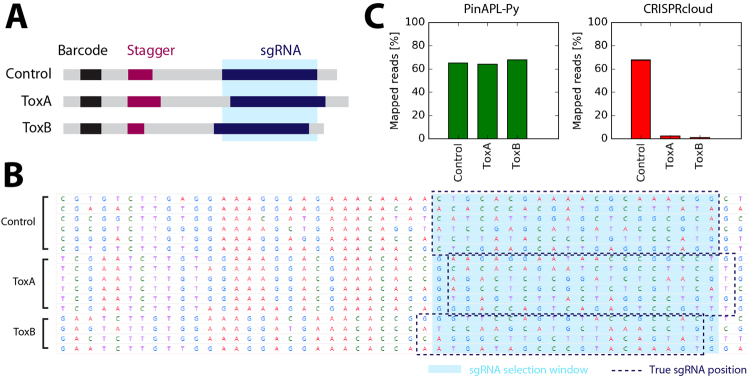
Figure 4.
Alignment of staggered reads. (A) Staggering introduces spacer sequences of variable length (purple) to cause shifts in the sequenced region, thereby enhancing nucleotide diversity per position when samples with different stagger lengths are pooled on the same sequencing lane. (B) Screenshot of CRISPRcloud’s read extraction page. The definition of the 20 bp sgRNA sequence (light blue highlight) needs to be defined for all samples at once. It, thus, misses the sgRNA region by +1 or −1 bp in the ToxA and the ToxB sample, respectively, due to the staggered read layout. (C) Since CRISPRcloud’s read identification does not to allow mismatches, read mapping fails in the corresponding samples.