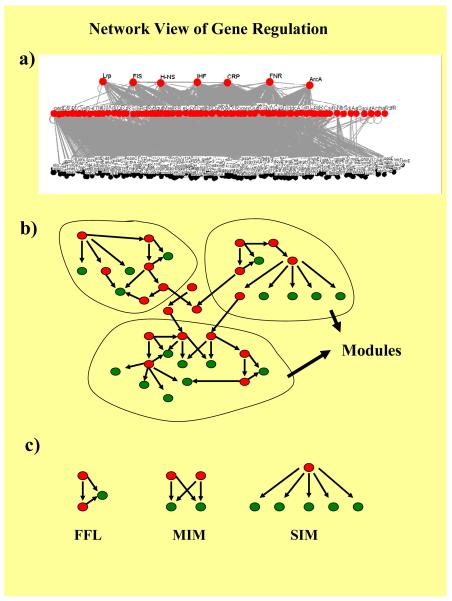
Fig. 2.
Network view of transcription regulation. Transcriptional regulatory interactions at a genomic level can be visualized as a network between TFs (shown in red) and target genes (shown in green). a) The transcriptional regulatory network is a multi-layer hierarchical modular structure without feedback regulation at the transcription level [38; 68], with the global regulators at the top of this layout and local TFs at the bottom, regulating a few genes. b) Modules are interconnected clusters which divide the network of transcriptional interactions into subnetworks. Modules have been identified using a variety of approaches [17; 38; 57] and have been found to be semi-independent in nature. Modules are in turn formed by one or more different types of network motifs. c) Motifs are patterns of interconnections which are overrepresented in transcriptional networks. Known transcriptional regulatory networks were found to have feed forward loops (FFLs), multiple input modules (MIMs) and SIMs, with each kind of motif playing a different role [1].