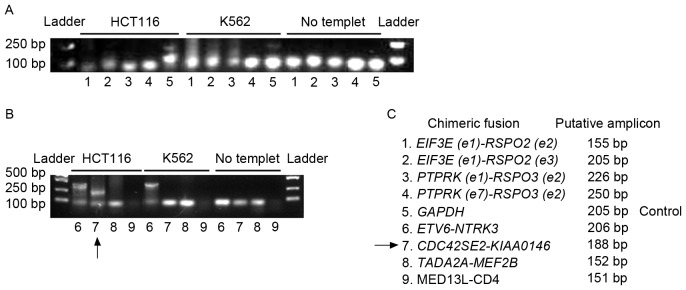
Figure 2.
Genomic fusion gene examination. Regular PCR was applied to determine the existence of fusion genes in the genome of HCT116 cells and K562 cells. (A) Detection for the existence of fusions 1 to 5. (B) Detection for the existence of fusions 6 to 9. PCR results were shown by agarose gel electrophoresis. ‘No template’ indicated the negative control of PCR, which was not supplied with template to preclude the contamination of the PCR system. GAPDH was used as a positive control for PCR amplification. The arrow indicates the amplicon of CDC42SE2-KIAAO146 from the HCT116 genome. The ETV6-NTRK3 fusion gene was detected in the HCT116 and K562 genome. (C) The code designation of fusion genes and the putative length of PCR amplicons were listed. PCR, polymerase chain reaction; GAPH, glyceraldehyde 3-phosphate dehydrogenase.