Abstract
Genome-wide association studies have identified multiple genetic polymorphisms associated with schizophrenia. These polymorphisms conform to a polygenic disease model in which multiple alleles cumulatively increase the risk of developing disease. Two genes linked to schizophrenia, DTNBP1 and MUTED, encode proteins that belong to the endosome-localized Biogenesis of Lysosome-related Organelles Complex-1 (BLOC-1). BLOC-1 plays a key role in endosomal trafficking, and as such has been found to regulate cell surface abundance of the D2 dopamine receptor (DRD2), the biogenesis and fusion of synaptic vesicles, and neurite outgrowth. These functions are pertinent to both neurodevelopment and synaptic transmission, processes tightly regulated by selective cell surface delivery of membrane proteins to and from endosomes. We propose that cellular processes, such as endosomal trafficking, act as convergence points where multiple small effects from polygenic genetic polymorphisms pileup to trigger schizophrenia.
Schizophrenia is a devastating mental illness that can lead to the inability to live independently within our society. Genetic factors and their interactions with the environment account for 80% of the susceptibility for disease emergence [1]. The identity of these genetic factors is the subject of intense scrutiny. Recent genome-wide studies suggest that the risk of developing schizophrenia is polygenic, with multiple common allelles each contributing a small effect [2]. Such studies have identified a plethora of polymorphisms associated with the illness. Moreover, the genes identified as associated with schizophrenia have markedly diverse functions; they include genes implicated in signal transduction, growth cone guidance, pre- and post-synaptic neurotransmission, and vesicular membrane protein trafficking [2–7].
Pathogenic hypotheses for schizophrenia have tended to emphasize individual genes of “interest” [1, 8]. Broadly, these hypotheses fall into two main categories: neurochemical or neurodevelopmental [8, 9]. The neurochemical hypotheses focus on alterations in neurotransmitter systems, such as dopamine, glutamate or GABA, and are substantiated by neurotransmitter abnormalities found in the brains of affected individuals, as well as the mechanism of action for pharmacological agents used in treating or inducing schizophrenia symptoms [9, 10]. The neurodevelopmental hypotheses encompass structural alterations in the brain of schizophrenic patients that emerge early in life [9, 11]. Here, we discuss studies by several groups that focus on the biogenesis of lysosome-related organelles complex-1 (BLOC-1) [12–14]. These studies implicate endosomal trafficking to and from the plasma membrane as a cellular mechanism that could link the neurochemical and neurodevelopmental hypotheses of schizophrenia.
Eight subunits constitute the BLOC-1 complex: dysbindin, muted, pallidin, cappuccino, snapin, BLOS1-3 [15]. This complex is present on transferrin-receptor positive endosomes, where it regulates membrane protein targeting to synaptic vesicles, lysosomes, and lysosome-related organelles [13, 16, 17]. BLOC-1 subunits are tightly bound, as evidenced by the observation that all of the dysbindin and pallidin protein in the brain copurify as a complex with the predicted molecular mass of the BLOC-1 complex [12]. Mice with genetic defects in BLOC-1 subunits further highlight the tight structural organization of BLOC-1 complex. Most null alleles of genes encoding BLOC-1 subunits lead to almost identical cell and organism phenotypes [18–21]. Among these phenotypes, the absence of any one of the BLOC-1 subunit proteins triggers the disappearance of all other BLOC-1 protein complex subunits [12, 18, 19, 22].
Two of the BLOC-1 complex subunits, dysbindin (encoded by DTNBP1) and muted (encoded by MUTED), have been associated with increased risk of schizophrenia [23, 24]. Several population genetic studies have replicated the association of DTNBP1 variants with schizophrenia [3, 8]. Importantly, dysbindin mRNA and protein abundance are decreased in the brain of schizophrenics [25, 26]. Based on these data, the hypothesis that BLOC-1 is implicated in the pathogenesis of schizophrenia leads to three predictions. First, BLOC-1-deficient mice should have neurological phenotypes consistent with the disease. Second, genetic polymorphisms in DTNBP1 associated with schizophrenia should trigger reduced levels of dysbindin in the brain of schizophrenic patients. Finally, brain tissue from schizophrenics with decreased dysbindin protein levels should also possess reduced levels in other BLOC-1 subunits, irrespective of whether these other BLOC-1 subunit genes were to carry disease-associated polymorphisms. The first prediction has been documented in dysbindin deficient mice, sandy, which display impaired social interactions and working memory both phenotypes consistent with schizophrenia [11, 27–29]. Similarly, DTNBP1 disease-associated polymorphisms decrease the levels of DTNBP1 transcripts in human cortex [30], thus supporting the second prediction. However, there is neither evidence to support the last prediction nor is there information about the developmental stage at which deficiencies in BLOC-1 function may impact the brain in schizophrenic patients.
Key observations of Ghiani et al. [12] suggest that BLOC-1 function may be required in the neonatal period, an observation consistent with the neurodevelopmental hypothesis of schizophrenia. First, they found that brain dysbindin and pallidin protein levels were the highest perinatally and declined in adulthood. Second, they described that loss of BLOC-1 in pallidin-null mice led to neurite outgrowth defects in primary cultured hippocampal neurons [12]. However, the effects of BLOC-1 extend beyond developmental processes. For instance, Iizuka et al. found that BLOC-1 decreases the cell-surface abundance and activity of the D2 dopamine receptor (DRD2). In neuronal cells where BLOC-1 was down-regulated by siRNA, the surface abundance of DRD2 and receptor-dependent signal transduction increased [31]. Intriguingly, DRD2, which encodes DRD2, is, like dysbindin, considered a candidate schizophrenia susceptibility gene [8]. Indeed, the D2 dopamine receptor plays a central role in the well-known dopamine hypothesis of schizophrenia, which postulates that increased dopaminergic neurotransmission is causative of disease [10]. In this context, increased surface levels of DRD2 receptors in neurons induced by defective expression of BLOC-1 would favor enhanced dopaminergic neurotransmission. Furthermore, genetic polymorphisms in the DRD2 gene may potentiate this enhanced dopaminergic neurotransmission. BLOC-1-dependent endosomal trafficking mechanisms in neurons may not limited to modulation of the DRD2 receptor, instead affecting the surface levels of diverse membrane proteins and thereby altering neuronal responsiveness to extracellular cues.
Although the precise endosomal processes affected by BLOC-1 deficiency are unclear, two non-exclusive mechanisms may contribute to the increased abundance of these proteins at the cell surface. Evidence indicates that BLOC-1 regulates the sorting of selected membrane proteins into vesicles either by itself or in association with the adaptor complex AP-3 [13, 16, 32, 33]. In fact, mouse models carrying null mutations in subunits of either BLOC-1 or ubiquitous AP-3 increase the content of specific synaptic vesicle proteins, most prominently VAMP7-TI, an endosomal SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) [13]. BLOC-1 also affects membrane fusion [12, 14]. Mice lacking dysbindin, and therefore the BLOC-1 complex, have slower quantal synaptic vesicle release and lower synaptic vesicles release probability [14]. This may be related, at least in part, to the ability of BLOC-1 complexes to bind and regulate the subcellular distribution of endosomal SNAREs [12, 13, 33], membrane proteins that control the selectivity of membrane fusion along the endocytic route and between endosomal-derived organelles and the cell surface, such as synaptic vesicles [34]. Regardless, either defective biogenesis or fusion of synaptic vesicles could decrease synaptic availability of neurotransmitter, a notion consistent with the glutamatergic and GABAergic neurochemical hypotheses of schizophrenia which postulate decreased activity of these neurotransmitter systems in schizophrenia brains [10].
An attractive aspect of an endosomal hypothesis is its ability to accommodate the polygenic nature of the genetic association data. For example, one-third of the 29 genes considered top scores for increased schizophrenia susceptibility in genetic association meta-analyses are either regulated by or participate in endosomal mechanisms [3] (see Table 1). Furthermore, the products of several genes identified as contributors to the risk of developing schizophrenia in recent genome wide studies are either regulated or participate in endosomal trafficking (Table 1). Consider two gene products from table 1: VMAT1 (encoded by SLC18A)[35], which is involved in presynaptic storage of dopamine in vesicles, and DRD2 (encoded by DRD2), a postsynaptic dopamine receptor. Recognizing how polymorphisms in these two genes fit to the dopaminergic hypothesis is straightforward. However, under the umbrella of an endosomal hypothesis we can understand how the products of several of the genes in table 1 could regulate dopaminergic neurotransmission. The subcellular distribution of VMAT1 and DRD2 could be under control of proteins involved in endosomal sorting and vesiculation encoded by genes such as DTNBP1 (dysbindin), CLTCL1 (clathrin isoform-1), and CENTG2 (ARF1 GTPase activating protein-1 (AGAP1)). The clathrin isoform encoded by the CLTCL1 gene is deleted from chromosome 22 of schizophrenia patients [4]. Clathrin and clathrin adaptor complexes orchestrate the formation of vesicles from multiple compartments in cells, including endosomes [36]. AGAP1, the protein encoded by CENTG2, regulates the recruitment of AP-3 to endosomal membranes, a necessary step in the formation of vesicles from endosomes [37]. Importantly, AGAP1 directly interacts with AP-3, which in turns associates with both clathrin and BLOC-1 to form vesicles from endosomes [37, 38]. Thus, under a polygenic model of schizophrenia, disease-associated alleles in SLC18A, for example, could have small and clinically silent individual effects in either the protein levels or the function of VMAT1. Minor VMAT1 phenotypes could be further potentiated and rendered clinically significant by the compounded effect of multiple other disease-associated alleles affecting, for example, mechanisms regulating dopamine receptor surface levels. Such genes will include those genes encoding BLOC-1 subunits and other molecules involved in endosomal sorting and vesiculation, such as clathrin, AP-3 and AGAP1.
Table 1. Schizophrenia Susceptibility Genes Related to Endocytic Trafficking.
Selected supporting references are listed with capital letters in the right hand column. The following studies are included SzGene study [3], the studies that identified microdeletions in chromosomes 22 (Chr22) and 15 (Chr15) [4, 7], genome wide association studies (GWAS) conducted on the Molecular Basis of Schizophrenia Study (MGS GWAS) [5], the Stefansson’s genome wide association study [6] and the International Schizophrenia consortium genome wide association studies (ISC GWAS) [2]. A) Mol Psychiatry. 2007;12:74–86. B) J Cell Biol. 1994;127:1419–33. C) Nat Rev Neurosci. 2007;8:413–26. D) Crit Rev Biochem Mol Biol. 2007;42:3–14. E) J Biol Chem. 2007;282:15778–89. F) Eur J Neurosci. 2006;24:1395–403 and Eur J Pharmacol. 2007;572:83–93. G) Adv Exp Med Biol. 2007;600:1–11. H) Mol Cell Neurosci. 2005;28:335–46. I) Mol Biol Cell. 2006;17:4027–38. J) Mol Biol Cell. 2004;15:3181–95. K) Mol Biol Cell. 2008;19:1942–51. L) J Biol Chem. 2004;279:51250–7. M) J Biol Chem. 2005;280:25769–79. N) Pflugers Arch. 2002;444:795–800. O) J Cell Biol. 2008;181:1179–93. P) J Cell Sci. 2006;119:1203–11. Q) Trends Neurosci. 2002;25:379–81. R) Biochim Biophys Acta. 2008;1782:99–108. S) Exp Cell Res. 2009;315:683–96. T) Science. 2009;325:213–7.
| GENE | Study | Gene Product Name | Regulated by or Involved in Endosome Traffic |
|---|---|---|---|
| DISC1 | SzGene | Disc1 | A |
| SLC18A1 | SzGene | VMAT1 | B |
| GRIN2B | SzGene | NMDA receptor subtype 2B | C |
| GABRB2 | SzGene | GABA A receptor, beta 2 | D |
| DRD1 | SzGene | Dopamine receptor 1 | E |
| DRD2 | SzGene | Dopamine receptor 2 | F |
| DRD4 | SzGene | Dopamine receptor 4 | Not explored |
| PLXNA2 | SzGene | Plexin A2 | G |
| NRG1 | SzGene & Stefansson GWAS | Neuregulin 1 | H |
| DTNBP1 | SzGene | Dysbindin | I |
| CLTCL1 | Chr 22 ISC | clathrin, heavy chain-like 1 | J |
| SEPT5 | Chr 22 ISC | Septin 5 | K |
| SCARF2 | Chr 22 ISC | scavenger receptor F 2 | L |
| SNAP29 | Chr 22 ISC | SNARE SNAP29 | M |
| P2RXL1 | Chr 22 ISC | purinergic receptor P2X 6 | N |
| CHRNA7 | Chr 15 ISC | cholinergic receptor, nicotinic, a7 | O |
| CENTG2 | MGS GWAS | AGAP1 | P |
| NTRK3 | MGS GWAS | TRKC | Q |
| ADPR2 | MGS GWAS | adiponectin receptor 2 | R |
| ERBB4 | MGS GWAS | Neuregulin receptor 4 | S |
| MHC locus | MGS, ISC, Stefansson GWAS | MHC gene products | T |
Endosomal trafficking controls cell surface receptor numbers in most cell types. Trafficking qualitatively and quantitatively affects signal transduction and cell surface composition in developing and adult organisms [39]. In neurons, endosomal trafficking mechanisms define pre- and post-synaptic composition and function by controlling the biogenesis of synaptic vesicles as well as the subcellular distribution of neurotransmitter transporters and receptors [40, 41]. Focusing on endosomal trafficking builds a conceptual bridge among traditional models of schizophrenia pathogenesis. Although we have focused on the BLOC-1 complex and its role in endosomal trafficking, analysis of present and future genetic data may define other fundamental cellular machineries or processes that could contribute to disease pathogenesis. We believe that focusing on cellular mechanisms rather than isolated genes of “interest” will facilitate the formulation of hypotheses with unique predictions amenable to exploration in the genomes and brain tissue of affected individuals.
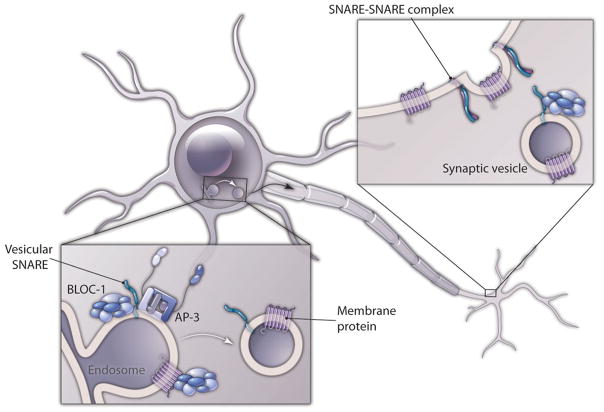
Figure 1. Putative events regulated by BLOC-1-dependent mechanisms in neurons.
Diagram depicts the biogenesis of lysosome-related organelles complex (BLOC)-1 possible sites of action. BLOC-1 modulates sorting of membrane proteins and SNAREs either alone or in conjunction with AP-3 at endosomes in the cell body impacting the composition of presynaptic secretory organelles. Additionally, BLOC-1 binds and may regulate SNARE-dependent membrane fusion at the nerve terminal.
Acknowledgments
This work was supported by grants from the National Institutes of Health to V.F. (NS42599) and T32 GM008367, National Institutes of Health, Training Program in Biochemistry, Cell, and Molecular Biology to P.V.R.
References
- 1.Tandon R, Keshavan MS, Nasrallah HA. Schizophrenia, “just the facts” what we know in 2008. 2. Epidemiology and etiology. Schizophrenia research. 2008;102:1–18. doi: 10.1016/j.schres.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 2.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P, Purcell Leader SM, Ruderfer DM, McQuillin A, Morris DW, O’Dushlaine CT, Corvin A, Holmans PA, Macgregor S, Gurling H, Blackwood DH, Craddock NJ, Gill M, Hultman CM, Kirov GK, Lichtenstein P, Muir WJ, Owen MJ, Pato CN, Scolnick EM, St Clair D, Sklar Leader P, Williams NM, Georgieva L, Nikolov I, Norton N, Williams H, Toncheva D, Milanova V, Thelander EF, Sullivan P, Kenny E, Quinn EM, Choudhury K, Datta S, Pimm J, Thirumalai S, Puri V, Krasucki R, Lawrence J, Quested D, Bass N, Crombie C, Fraser G, Leh Kuan S, Walker N, McGhee KA, Pickard B, Malloy P, Maclean AW, Van Beck M, Pato MT, Medeiros H, Middleton F, Carvalho C, Morley C, Fanous A, Conti D, Knowles JA, Paz Ferreira C, Macedo A, Helena Azevedo M, Kirby AN, Ferreira MA, Daly MJ, Chambert K, Kuruvilla F, Gabriel SB, Ardlie K, Moran JL. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009 doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, Tanzi RE, Bertram L. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nature genetics. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 4.Consortium IS. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, Dudbridge F, Holmans PA, Whittemore AS, Mowry BJ, Olincy A, Amin F, Cloninger CR, Silverman JM, Buccola NG, Byerley WF, Black DW, Crowe RR, Oksenberg JR, Mirel DB, Kendler KS, Freedman R, Gejman PV. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009 doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietilainen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Borglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Bottcher Y, Olesen J, Breuer R, Moller HJ, Giegling I, Rasmussen HB, Timm S, Mattheisen M, Bitter I, Rethelyi JM, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, Gulcher JR, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA, Group, Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, de Frutos R, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Lim Yoon J, Shianna KV, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jonsson EG, Terenius L, Agartz I, Petursson H, Nothen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, St Clair D, Goldstein DB, Stefansson K, Collier DA, Kahn RS, Linszen DH, van Os J, Wiersma D, Bruggeman R, Cahn W, de Haan L, Krabbendam L, Myin-Germeys I. Common variants conferring risk of schizophrenia. Nature. 2009 doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Moller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, Di Forti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Muhleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nothen MM, Peltonen L, Collier DA, St Clair D, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross CA, Margolis RL, Reading SA, Pletnikov M, Coyle JT. Neurobiology of schizophrenia. Neuron. 2006;52:139–153. doi: 10.1016/j.neuron.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 9.Lieberman JA, Stroup TS, Perkins DO American Psychiatric Publishing. The American Psychiatric Publishing textbook of schizophrenia. 1. American Psychiatric Pub; Washington, DC: 2006. [Google Scholar]
- 10.Keshavan MS, Tandon R, Boutros NN, Nasrallah HA. Schizophrenia, “just the facts”: what we know in 2008 Part 3: neurobiology. Schizophrenia research. 2008;106:89–107. doi: 10.1016/j.schres.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 11.Jarskog LF, Miyamoto S, Lieberman JA. Schizophrenia: new pathological insights and therapies. Annu Rev Med. 2007;58:49–61. doi: 10.1146/annurev.med.58.060904.084114. [DOI] [PubMed] [Google Scholar]
- 12.Ghiani CA, Starcevic M, Rodriguez-Fernandez IA, Nazarian R, Cheli VT, Chan LN, Malvar JS, de Vellis J, Sabatti C, Dell’angelica EC. The dysbindin-containing complex (BLOC-1) in brain: developmental regulation, interaction with SNARE proteins and role in neurite outgrowth. Mol Psychiatry. 2009 doi: 10.1038/mp.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newell-Litwa K, Salazar G, Smith Y, Faundez V. Roles of BLOC-1 and AP-3 Complexes in Cargo Sorting to Synaptic Vesicles. Molecular biology of the cell. 2009;20:1441–1453. doi: 10.1091/mbc.E08-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen XW, Feng YQ, Hao CJ, Guo XL, He X, Zhou ZY, Guo N, Huang HP, Xiong W, Zheng H, Zuo PL, Zhang CX, Li W, Zhou Z. DTNBP1, a schizophrenia susceptibility gene, affects kinetics of transmitter release. The Journal of cell biology. 2008;181:791–801. doi: 10.1083/jcb.200711021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Di Pietro SM, Dell’Angelica EC. The cell biology of Hermansky-Pudlak syndrome: recent advances. Traffic. 2005;6:525–533. doi: 10.1111/j.1600-0854.2005.00299.x. [DOI] [PubMed] [Google Scholar]
- 16.Di Pietro SM, Falcon-Perez JM, Tenza D, Setty SR, Marks MS, Raposo G, Dell’angelica EC. BLOC-1 Interacts with BLOC-2 and the AP-3 Complex to Facilitate Protein Trafficking on Endosomes. Molecular biology of the cell. 2006;17:4027–4038. doi: 10.1091/mbc.E06-05-0379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Setty SR, Tenza D, Truschel ST, Chou E, Sviderskaya EV, Theos AC, Lamoreux ML, Di Pietro SM, Starcevic M, Bennett DC, Dell’Angelica EC, Raposo G, Marks MS. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Molecular biology of the cell. 2007;18:768–780. doi: 10.1091/mbc.E06-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciciotte SL, Gwynn B, Moriyama K, Huizing M, Gahl WA, Bonifacino JS, Peters LL. Cappuccino, a mouse model of Hermansky-Pudlak syndrome, encodes a novel protein that is part of the pallidin-muted complex (BLOC-1) Blood. 2003;101:4402–4407. doi: 10.1182/blood-2003-01-0020. [DOI] [PubMed] [Google Scholar]
- 19.Li W, Zhang Q, Oiso N, Novak EK, Gautam R, O’Brien EP, Tinsley CL, Blake DJ, Spritz RA, Copeland NG, Jenkins NA, Amato D, Roe BA, Starcevic M, Dell’Angelica EC, Elliott RW, Mishra V, Kingsmore SF, Paylor RE, Swank RT. Hermansky-Pudlak syndrome type 7 (HPS-7) results from mutant dysbindin, a member of the biogenesis of lysosome-related organelles complex 1 (BLOC-1) Nature genetics. 2003;35:84–89. doi: 10.1038/ng1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starcevic M, Dell’Angelica EC. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1) J Biol Chem. 2004;279:28393–28401. doi: 10.1074/jbc.M402513200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Q, Li W, Novak EK, Karim A, Mishra VS, Kingsmore SF, Roe BA, Suzuki T, Swank RT. The gene for the muted (mu) mouse, a model for Hermansky-Pudlak syndrome, defines a novel protein which regulates vesicle trafficking. Hum Mol Genet. 2002;11:697–706. doi: 10.1093/hmg/11.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falcon-Perez JM, Starcevic M, Gautam R, Dell’Angelica EC. BLOC-1, a novel complex containing the pallidin and muted proteins involved in the biogenesis of melanosomes and platelet-dense granules. J Biol Chem. 2002;277:28191–28199. doi: 10.1074/jbc.M204011200. [DOI] [PubMed] [Google Scholar]
- 23.Guo AY, Sun J, Riley BP, Thiselton DL, Kendler KS, Zhao Z. The dystrobrevin-binding protein 1 gene: features and networks. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Straub RE, Mayhew M, Vakkalanka R, Kolachana B, Goldberg T, Egan M, Weinberger DR. MUTED (6p24.3), a Protein That Binds to Dysbindin (DTNBP1, 6p22.3), Is Strongly Associated with Schizophrenia and Exhibits Statistical Epistasis with COMT. Neuropsychopharmacology. 2005;30:S1–S270. [Google Scholar]
- 25.Talbot K, Eidem WL, Tinsley CL, Benson MA, Thompson EW, Smith RJ, Hahn CG, Siegel SJ, Trojanowski JQ, Gur RE, Blake DJ, Arnold SE. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J Clin Invest. 2004;113:1353–1363. doi: 10.1172/JCI20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weickert CS, Rothmond DA, Hyde TM, Kleinman JE, Straub RE. Reduced DTNBP1 (dysbindin-1) mRNA in the hippocampal formation of schizophrenia patients. Schizophrenia research. 2008;98:105–110. doi: 10.1016/j.schres.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhardwaj SK, Baharnoori M, Sharif-Askari B, Kamath A, Williams S, Srivastava LK. Behavioral characterization of dysbindin-1 deficient sandy mice. Behav Brain Res. 2009;197:435–441. doi: 10.1016/j.bbr.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Cox MM, Tucker AM, Tang J, Talbot K, Richer DC, Yeh L, Arnold SE. Neurobehavioral abnormalities in the dysbindin-1 mutant, sandy, on a C57BL/6J genetic background. Genes Brain Behav. 2009;8:390–397. doi: 10.1111/j.1601-183X.2009.00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jentsch JD, Trantham-Davidson H, Jairl C, Tinsley M, Cannon TD, Lavin A. Dysbindin Modulates Prefrontal Cortical Glutamatergic Circuits and Working Memory Function in Mice. Neuropsychopharmacology. 2009 doi: 10.1038/npp.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bray NJ, Preece A, Williams NM, Moskvina V, Buckland PR, Owen MJ, O’Donovan MC. Haplotypes at the dystrobrevin binding protein 1 (DTNBP1) gene locus mediate risk for schizophrenia through reduced DTNBP1 expression. Hum Mol Genet. 2005;14:1947–1954. doi: 10.1093/hmg/ddi199. [DOI] [PubMed] [Google Scholar]
- 31.Iizuka Y, Sei Y, Weinberger DR, Straub RE. Evidence that the BLOC-1 protein dysbindin modulates dopamine D2 receptor internalization and signaling but not D1 internalization. J Neurosci. 2007;27:12390–12395. doi: 10.1523/JNEUROSCI.1689-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Setty SR, Tenza D, Sviderskaya EV, Bennett DC, Raposo G, Marks MS. Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature. 2008 doi: 10.1038/nature07163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salazar G, Craige B, Styers ML, Newell-Litwa KA, Doucette MM, Wainer BH, Falcon-Perez JM, Dell’angelica EC, Peden AA, Werner E, Faundez V. BLOC-1 Complex Deficiency Alters the Targeting of Adaptor Protein Complex-3 Cargoes. Molecular biology of the cell. 2006;17:4014–4026. doi: 10.1091/mbc.E06-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaudhry FA, Edwards RH, Fonnum F. Vesicular neurotransmitter transporters as targets for endogenous and exogenous toxic substances. Annu Rev Pharmacol Toxicol. 2008;48:277–301. doi: 10.1146/annurev.pharmtox.46.120604.141146. [DOI] [PubMed] [Google Scholar]
- 36.Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 37.Nie Z, Boehm M, Boja ES, Vass WC, Bonifacino JS, Fales HM, Randazzo PA. Specific regulation of the adaptor protein complex AP-3 by the Arf GAP AGAP1. Dev Cell. 2003;5:513–521. doi: 10.1016/s1534-5807(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 38.Salazar G, Zlatic S, Craige B, Peden AA, Pohl J, Faundez V. Hermansky-Pudlak syndrome protein complexes associate with phosphatidylinositol 4-kinase type II alpha in neuronal and non-neuronal cells. J Biol Chem. 2009;284:1790–1802. doi: 10.1074/jbc.M805991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Zastrow M, Sorkin A. Signaling on the endocytic pathway. Curr Opin Cell Biol. 2007;19:436–445. doi: 10.1016/j.ceb.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy MJ, Ehlers MD. Organelles and trafficking machinery for postsynaptic plasticity. Annu Rev Neurosci. 2006;29:325–362. doi: 10.1146/annurev.neuro.29.051605.112808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murthy VN, De Camilli P. Cell biology of the presynaptic terminal. Annu Rev Neurosci. 2003;26:701–728. doi: 10.1146/annurev.neuro.26.041002.131445. [DOI] [PubMed] [Google Scholar]