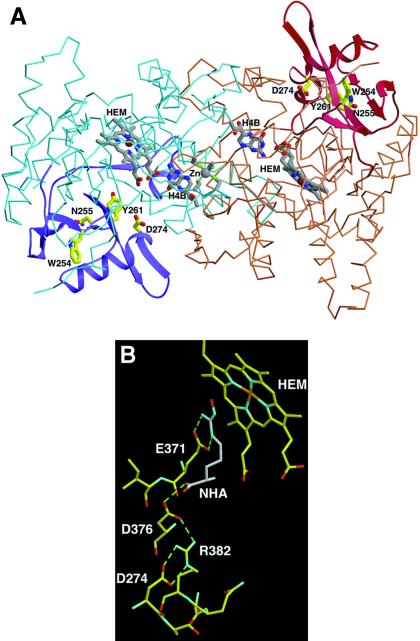
Figure 6.
(A) The subdomain encoded by exons 8 and 9 (residues 236–329 of mouse iNOS, corresponding to residues 242–335 of human iNOS) in the crystal structure of the H4B-bound mouse Δ 65 iNOSox dimer (PDB ID code 1NOD). Mouse iNOS residues corresponding to the human residues mutated in this study are labeled. iNOSox dimer (blue and brown subunits), heme (gray), H4B, and zinc (Zn) are shown as an α-carbon trace. The subdomain encoded by exons 8 and 9 (ribbons of darker color, purple and maroon) forms a wing of the “winged β-sheet” that structures the heme pocket and stabilizes the subunit fold. W254 (W260 in human iNOS), N255 (N261, human), Y261 (Y267, human), and D274 (D280, human) are critical for stabilizing the structural integrity of the subdomain and active center (yellow side chains). (B) Specific interaction of D274 (human D280) in the active-center channel and its role in stabilizing the substrate-binding site. Residue D274 forms a hydrogen-bonding network with Arg (R) 382 and residues D376 and E371 on the substrate-binding helix α7a that interact directly with substrate l-Arg and intermediate l-NOHA (shown in gray). Thus, mutation of D274 (human D280) removes important buttressing hydrogen bonds to conserved residues that stabilize substrate binding and catalytic activity.