Abstract
We review RNA interference (RNAi) of insect pests and its potential for implementing sterile insect technique (SIT)‐related control. The molecular mechanisms that support RNAi in pest species are reviewed in detail, drawing on literature from a range of species including Drosophila melanogaster Meigen and Homo sapiens L. The underlying genes that enable RNAi are generally conserved across taxa, although variance exists in both their form and function. RNAi represents a plausible, non‐GM system for targeting populations of insects for control purposes, if RNAi effector molecules can be delivered environmentally (eRNAi). We consider studies of eRNAi from across several insect orders and review to what extent taxonomy, genetics, and differing methods of double‐stranded (ds) RNA synthesis and delivery can influence the efficiency of gene knockdown. Several factors, including the secondary structure of the target mRNA and the specific nucleotide sequence of dsRNA effector molecules, can affect the potency of eRNAi. However, taxonomic relationships between insects cannot be used to reliably forecast the efficiency of an eRNAi response. The mechanisms by which insects acquire dsRNA from their environment require further research, but the evidence to date suggests that endocytosis and transport channels both play key roles. Delivery of RNA molecules packaged in intermediary carriers such as bacteria or nanoparticles may facilitate their entry into and through the gut, and enable the evasion of host defence systems, such as toxic pH, that would otherwise attenuate the potential for RNAi.
Keywords: insect control, environmental RNAi, SIT, non‐GM pest control, double‐stranded RNA
RNAi and the sterile insect technique (SIT)
Established methods of insect control are under continual review and development in order to keep track of new knowledge, changing legislation, regulatory concerns, and the maintenance of efficacy (e.g., in the face of increased resistance to pesticides) (Gross, 2013; Tabashnik et al., 2014). In this context, the development of new methods for insect control is of key importance and there has been intense interest in the utility of gene silencing methods induced by RNA interference (RNAi). RNAi can induce mortality (Yang & Han, 2014; Cao et al., 2015; Abd El Halim et al., 2016; Christiaens et al., 2016; Hu et al., 2016; Malik et al., 2016), create beneficial phenotypes for insect control (Salvemini et al., 2009; Shukla & Palli, 2012; Peng et al., 2015; Yu et al., 2016), and prevent pesticide resistance in insect pests (Figueira‐Mansur et al., 2013; Guo et al., 2015; Wei et al., 2015; Bona et al., 2016; Sandoval‐Mojica & Scharf, 2016). Therefore, the potential for RNAi as a basis for future pest management strategies holds great promise (Huvenne & Smagghe, 2010; Gu & Knipple, 2013; Scott et al., 2013; Baum & Roberts, 2014; Kim et al., 2015). The purpose of this review is to summarize the mechanisms by which gene silencing is achieved, describe the ways in which it is currently being used, and to explore the many factors that affect the efficacy of RNAi in this context.
RNAi can be used to achieve knock‐down of the level of gene expression in specific target genes. This is done via the introduction, by various means, of double‐stranded RNA (dsRNA) into the cells of the target species (Fire et al., 1998). The evidence suggests that RNAi is facilitated by the canonical small interfering RNA (siRNA) pathway, which results in mRNA degradation (Figure 1). In our review of the mechanisms of RNAi in pest insects we draw strongly from the well‐described canonical siRNA pathway in Drosophila melanogaster Meigen and Homo sapiens L.
Figure 1.
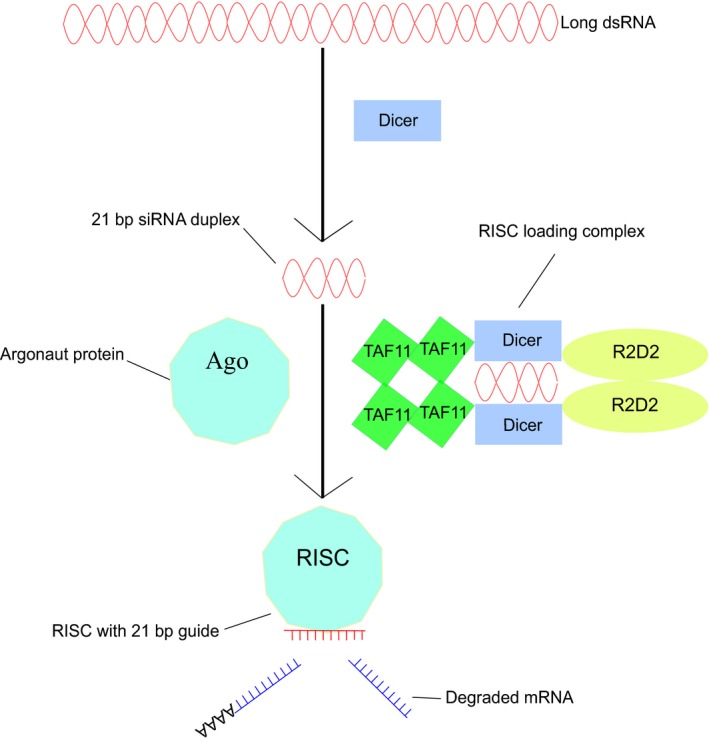
The canonical siRNA pathway. Cytoplasmic long double‐stranded RNAs are processed into 21‐bp duplex siRNAs by Dicer endonucleases. Dicer then complexes with various molecules to form a RISC loading complex (RLC) (the proposed RLC variant found in Drosophila melanogaster is shown here; Liang et al., 2015). The RLC introduces siRNA to an Argonaute protein, which degrades a single ‘passenger’ strand of the duplex, whilst binding its cognate partner to form an RNA induced silencing complex (RISC). The RISC then utilizes the nucleotide sequence of the bound ‘guide’ strand to scan cellular mRNAs, which it targets for knockdown via degradation.
It has been increasingly realized that a classic method of insect control, the sterile insect technique (SIT) (Knipling, 1955) could, in principle, be implemented through RNAi (Whyard et al., 2015). The SIT relies upon the production of large numbers of sterile insects for release (usually males) that subsequently mate with wild individuals, resulting in sterile matings and a reduction in the pest population size (Knipling, 1998; Krafsur, 1998). The key to SIT is the effective production of large numbers of sterile individuals. This crucial step is also a potential weakness of the approach. For example, the induction of sterility through irradiation results in well‐documented costs to insect performance, and hence control potential (Hooper, 1972; Toledo et al., 2004; Guerfali et al., 2011). Newer developments based on SIT that avoid irradiation, e.g., genetically engineered ‘self‐limiting’ insects (Thomas et al., 2000), can be highly effective (Harris et al., 2011; Carvalho et al., 2015; Gorman et al., 2016) but rely upon the release of genetically engineered insects, which may not be possible in all countries.
The principles by which RNAi might offer an alternative route for the induction of sterility, as well as other potentially useful manipulations for insect control, were recently investigated in a study using Aedes aegypti (L.) (Whyard et al., 2015). The scenario envisaged by Whyard et al. (2015) requires knockdown of at least two genes in the target insects. First, females would be targeted through silencing of a gene in the sexual differentiation cascade to turn them into pseudomales, i.e., genetic females which are phenotypically male (Pane et al., 2002; Salvemini et al., 2009; Shukla & Palli, 2012; Liu et al., 2015). Next, genes that could induce male (and pseudomale) sterility would be targeted in order to produce a 100% sterile male release cohort (Whyard et al., 2015). However, two equally important conditions must be met before this technique can be applied in the field, as described below.
The primary condition of RNAi‐based SIT is that the sex reversal target must reliably produce a male‐only cohort. There are clear benefits of releasing only one sex in SIT programmes, for example it can avoid both assortative mating between released insects and any pest‐related damage caused by females. The second condition is to ensure that silencing of neither the sex reversal nor the sterility target unduly reduces insect performance. Evidence suggests that these conditions can be met, although further supporting research is required.
Through RNAi of transformer‐2, Salvemini et al. (2009) were able to produce a Ceratitis capitata (Wiedemann) cohort which was 95.6% phenotypically male. Karyotypic analysis of phenotypically male flies (n = 20) demonstrated that they were 55% genetically female. Most importantly, pseudomales were observed completing male‐specific courtship rituals, which should allow them to attract and copulate with females (Briceño & Eberhard, 2003). Gabrieli et al. (2016) report that RNAi of innexin‐5 in C. capitata produced spermless, sterile males. Spermless males remained sexually competitive with wild‐type rivals and were able to induce similar post‐mating responses. It is possible that simultaneous RNAi of transformer‐2 and innexin‐5 (or conserved homologous genes in diverse species) could produce a male‐only, sterile cohort that could be used for SIT. However, it is important to note that simultaneous gene silencing is unpredictable (Table 2) and that Gabrieli et al. (2016) and Salvemini et al. (2009) microinjected insect eggs with dsRNA, a technique that is incompatible with large‐scale SIT.
Table 2.
Practical considerations when designing eRNAi‐based SIT strategies
| eRNAi is dose‐dependent. | The potency of gene silencing correlates with dsRNA concentration and period of exposure (Zhou et al., 2008; Tian et al., 2009, 2015; Singh et al., 2013; Asokan et al., 2014; Yu et al., 2014; Li et al., 2015b; Ulrich et al., 2015; Whyard et al., 2015; Rebijith et al., 2016). A period of latency between dsRNA administration and gene silencing is common in larvae and adults, which is consistent with a threshold effect. Reported latency periods include: 12 h in Helicoverpa armigera (Tian et al., 2015) and Aedes aegypti (Coy et al., 2012), 24 h in Mamestra configurata (Toprak et al., 2013) and Bactrocera dorsalis (Zheng et al., 2015), 7 days in Spodoptera exigua (Tian et al., 2009), and 12 days in Solenopsis invicta (Choi et al., 2012). It is of note that Choi et al. (2012) did not feed dsRNA to insects directly but via a secondary worker individual, and this may have diluted dsRNA somewhat leading to an extended latency period. |
| Taxonomy cannot be used to reliably predict sensitivity to eRNAi or the latency period between dsRNA uptake and gene silencing. | Equivalent eRNAi methods can produce disparate results even when different biotypes (Li et al., 2015a), or subpopulations (Sugahara et al., 2017) of the same species are targeted. Within the Lepidoptera, H. armigera is capable of eliciting a robust eRNAi response (Xiong et al., 2013), but Spodoptera frugiperda is recalcitrant to eRNAi (Ivashuta et al., 2015). Working on hemipteran and dipteran models, Coleman et al. (2015) and Yi et al. (2014) observed a 4‐day period of latency between dsRNA administration and gene silencing. Another dipteran (A. aegypti) exhibited gene silencing after 12 h of feeding with dsRNA in solution (Coy et al., 2012). These discrepancies may be eliminated if equivalent feeding protocols are used. |
| The sensitivity of a gene to RNAi has not been fully assessed unless the entire mRNA molecule has been targeted for knockdown. | When all variables remain constant, variation in RNAi potency is likely to be due to regional susceptibility of mRNAs to cleavage. The strength of gene knockdown can vary greatly when multiple genes are targeted in the same insect using identical methods (Pridgeon et al., 2008; Li et al., 2011; Singh et al., 2013; Toprak et al., 2013; Killiny et al., 2014; Taracena et al., 2015). |
| Insects may become more tolerant to dsRNA with aging. | A diminution in the efficiency of RNAi with age has been suggested by Tian et al. (2015) and is supported by evidence that silencing appears more efficient in neonates than in late stage larvae (Zhu et al., 2011; Toprak et al., 2013). Furthermore, Coleman et al.'s (2015) observation that silencing is longer lived in nymphs than in adults suggests that fully developed insects are less sensitive to dsRNA. |
| Optimum eRNAi delivery methods must be determined by trial and error in most insect species. | Yang & Han (2014) found that feeding H. armigera with transgenic bacteria induced more efficient eRNAi than feeding with naked dsRNA. However, naked dsRNA elicited more robust gene silencing than did bacterial feeding in B. dorsalis (Li et al., 2011). Zhu et al. (2011) report that three of five genes were knocked down more efficiently using a bacterial system in Leptinotarsa decemlineata, but that the remaining two were more efficiently silenced by naked dsRNA. |
| When inducing eRNAi the capacity for systemic RNAi is critical if target genes lie beyond gut tissue. | The systemic RNAi capacity of various insects has been assessed by targeting chitin synthase genes specific to the exoskeleton (Tian et al., 2009; Zhang et al., 2010; Singh et al., 2013). Other examples of studies that targeted genes distal to gut tissue include the silencing of ebony in Diabrotica virgifera vigifera (Miyata et al., 2014) and Rhodnius heme binding protein (RHBP) in Rhodnius prolixus (Taracena et al., 2015). Ivashuta et al. (2015) suggest that systemic RNAi in D. v. virgifera is facilitated by transport of dsRNAs of >60 bp long. |
| Parental RNAi requires further analysis to determine whether it can be effective for SIT. | A robust systemic RNAi response enables the silencing of genes in germ cells (parental RNAi; pRNAi). When germ cells are affected by pRNAi, gene expression can be limited in zygotes and developing insects (Zwier et al., 2012; Paim et al., 2013; Coleman et al., 2015; Khajuria et al., 2015). A simple application of pRNAi in pest management would be to reduce future insect populations via embryonic lethal gene silencing (Khajuria et al., 2015). For use with SIT, sexual differentiation genes could be targeted in the mothers of target insects (Shukla & Palli, 2012). dsRNA delivery methods may drastically affect the potency of pRNAi. Zheng et al. (2015) report that eRNAi silencing of sex peptide receptor in B. dorsalis limited eclosion rates of their progeny, whereas Peng et al. (2015) describe that silencing the transformer gene by microinjection in this species has no effect on progeny. The disparity in pRNAi efficiency between these delivery methods might be due to the fact that eRNAi would have consistently supplied flies with dsRNA during the development of germ cells, whereas expression of transformer would have been only transiently reduced by microinjection. |
| Insects may become less sensitive to dsRNA over time. | Working with B. dorsalis, Li et al. (2015b) demonstrated that eRNAi potency was reduced following a series of exposures to dsRNA. The effect was dose‐dependent, was not gene specific, and lasted for up to 20 days following primary exposure. Refractoriness only occurred following targeting of endogenous genes, which suggests a role in immune priming. Li et al. (2015b) suggest that flies may become refractory to dsRNA when genes that mediate endocytosis are downregulated. Bactrocera dorsalis has also been reported to upregulate the expression of target genes following exposure to dsRNA (Li et al., 2011). Therefore, reduced eRNAi potency may be due to synergistic overexpression of target genes along with downregulation of endocytic mediators. |
| Endocytic pathways and SID transport proteins may work synergistically in eRNAi. | Both endocytosis and SID mediated dsRNA transport facilitate eRNAi in L. decemlineata (Cappelle et al., 2016). |
| Insects may become more sensitive to eRNAi if dsRNA is vectored in nanoparticles. The performance of various nanoparticle technologies for use with RNAi is reviewed in Liao et al. (2016). | Nanoparticle‐based delivery of dsRNA may serve dual purposes: (1) enhancing passage of dsRNA across the gut, and (2) prolonging the effect of RNAi via slow release of dsRNA. Whyard et al. (2009) utilized Lipofectamine 2000 and Cellfectin liposomal nanoparticles to successfully vector dsRNA to Drosophila melanogaster, even though this species is reported as eRNAi incompetent. Liposomal vectoring achieved ca. 50% gene knockdown. Recently, Drosophila suzukii (Matsumura) has also been reported as recalcitrant to feeding with naked dsRNA, but sensitive if molecules are vectored in liposomes (Taning et al., 2016). Chitosan nanoparticles can vector molecules to Anopheles gambiae and A. aegypti (Zhang et al., 2010, 2015b; Mysore et al., 2015), although mosquito larvae (Figueira‐Mansur et al., 2013; Singh et al., 2013; Whyard et al., 2015) and adults (Pridgeon et al., 2008; Coy et al., 2012) also demonstrate eRNAi when exposed to naked dsRNA. Potent eRNAi has been demonstrated in A. aegypti larvae using carbon quantum dot (CQD) nanoparticles (Das et al., 2015). |
| Data regarding the simultaneous knockdown of genes via administration of multiple dsRNAs are conflicting. | Simultaneous silencing of genes has been reported to enhance the potency of RNAi in A. aegypti using a bacterial feeding approach (Whyard et al., 2015). A plant feeding study of Myzus persicae and Bactericera cockerelli also suggested that targeting genes simultaneously may induce a synergistic effect (Tzin et al., 2015). Zhang et al. (2015a) and Ulrich et al. (2015) report simultaneous silencing in coleopteran models actually dilutes the potency of RNAi. When feeding combinations of dsRNAs to D. suzukii, Taning et al. (2016) found the potency of RNAi was enhanced for some target gene combinations but not others. |
Microinjection of dsRNA has been demonstrated to induce RNAi in several insects (Paim et al., 2013; Peng et al., 2015; Xue et al., 2015; Yu et al., 2016). However, SIT programmes may require the production and release of up to a billion insects per week (Alphey et al., 2010) and injection techniques cannot be used to treat insects in such numbers. Therefore, RNAi may provide a useful tool for implementing SIT if gene silencing can be induced via environmental dsRNA (eRNAi) (Whangbo & Hunter, 2008).
Cell autonomous RNAi defines gene silencing in response to intracellular dsRNA of experimental or viral origin. Non‐cell autonomous RNAi defines gene silencing in response to an extracellular signal, and is further divided into systemic RNAi or eRNAi based on the nature of that signal. eRNAi describes gene silencing in response to proximal dsRNA molecules, whereas systemic describes RNAi gene silencing in response to a signal received from a proximal cell. Therefore, both non‐cell autonomous RNAi and eRNAi occur in a primary cell in direct response to dsRNA, whereas systemic RNAi is initiated in a secondary cell in response to an as yet undefined signal received from a primary cell (Figure 2). eRNAi can be achieved via the introduction of dsRNAs via food (Asokan et al., 2014; Coleman et al., 2015; Li et al., 2015b; Sandoval‐Mojica & Scharf, 2016) or through topical delivery (Toprak et al., 2013; Whyard et al., 2015). For reasons that are as yet not entirely clear, the capacity of insects to express systemic RNAi and eRNAi varies both within and between species (Baum & Roberts, 2014; Li et al., 2015a; Shukla et al., 2016; Sugahara et al., 2017).
Figure 2.
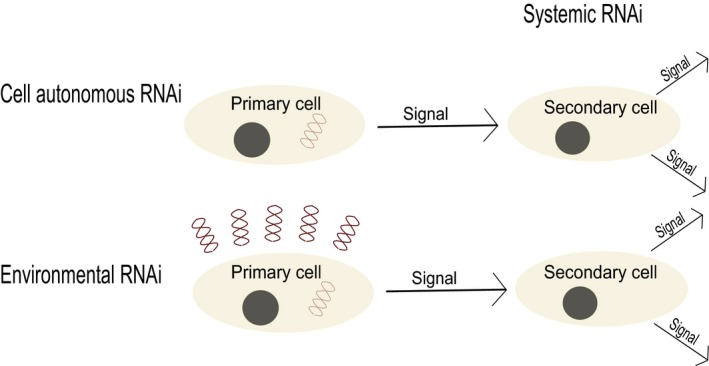
Categories of RNAi response. Cell autonomous RNAi is gene silencing in response to cytoplasmic dsRNA of viral or experimental origin. Non‐cell autonomous RNAi occurs in response to an extracellular signal, and is subcategorized by the origin of that signal as either environmental (eRNAi), or systematic RNAi. eRNAi occurs when a cell takes up environmental dsRNA molecules and elicits a gene silencing response. Systemic RNAi is initiated in a secondary cell when a silencing signal is received from a primary cell. Systemic RNAi can be a by‐product of either non‐cell autonomous RNAi or eRNAi in a primary cell, and if the secondary cell further propagates the signal, this can induce global gene silencing.
Many factors affect the efficiency of gene silencing induced by eRNAi. Some are intrinsic properties of the insects themselves (genetic differences, feeding habits, etc.), but others correspond to the nature of dsRNA effector molecules and their state at the point of encounter/entry to the host. In this review, we first describe the mechanisms of RNAi in detail, highlight examples of its use in different pest species, and in the concluding section consider the factors affecting eRNAi, in an attempt to discover whether there are emergent properties that might be useful in the planning of SIT strategies.
Mechanism of RNAi
RNAi is facilitated through the canonical siRNA pathway, culminating in the degradation of target mRNA
Small interfering RNAs (siRNAs) are short (ca. 21 nt) double‐stranded RNA molecules that are cleaved from long, cytoplasmic dsRNA transcripts (Figure 3). siRNAs belong to a large family of small non‐coding RNAs (ncRNAs) that facilitate different modes of gene silencing. ncRNA species include small interfering RNAs (siRNA), microRNAs (miRNA), PIWI‐interacting RNAs (piRNA), trans‐acting RNAs (tasiRNAs), repeat‐associated RNAs (rasiRNAs), and small‐scan RNAs (scnRNAs) (Kuramochi‐Miyagawa et al., 2001; Kim et al., 2009, 2015). While the origin and function of each ncRNA species is distinct (Bartel, 2004; Gasciolli et al., 2005; Babiarz et al., 2008; Kim et al., 2009) their actions are facilitated by homologous molecular mechanisms.
Figure 3.
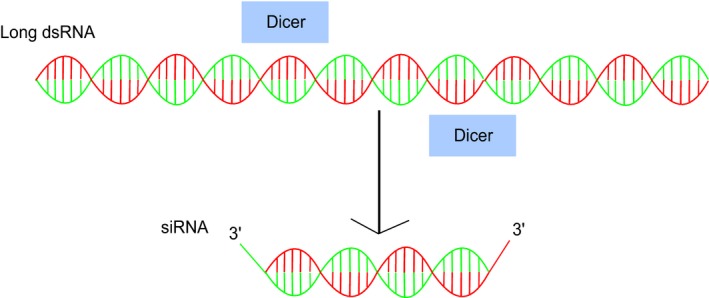
siRNA biogenesis. 21‐nt siRNA duplexes are cleaved from long, cytoplasmic dsRNA molecules by Dicer endonucleases. Two cuts are carried out by discrete Dicer RNAase III motifs, leaving short 3′ overhangs on each strand (Tomari & Zamore, 2005).
ncRNAs can only initiate gene silencing when bound to an Argonaute protein as part of an RNA‐induced silencing complex (RISC). When assembled in a RISC, exo‐siRNAs target viral mRNAs for knockdown as part of an immune response (Lan et al., 2016a,b). In contrast, endo‐siRNAs target endogenously transcribed mRNAs in order to achieve gene regulation (Babiarz et al., 2008; Okamura et al., 2008). exo‐siRNAs function through the canonical siRNA pathway, inducing cleavage of target mRNAs (Elbashir et al., 2001a; Song et al., 2004), while endo‐siRNAs inhibit the translation of target molecules (Hannon, 2002).
The canonical siRNA pathway requires the nucleotide sequences of siRNA molecules and their intended mRNA targets to exhibit almost perfect complementarity (Joseph & Osman, 2012). Imperfect homology may result in a mode of gene silencing other than mRNA cleavage (such as translational repression; Hu et al., 2010), which is associated with other ncRNA pathways. Perfect sequence homology is achievable in RNAi, as the target mRNA can usually be used to design effector dsRNA molecules with perfect matching. Therefore, the predominant mechanism of gene silencing induced by RNAi is mRNA degradation.
siRNA biogenesis: Dicer
siRNAs are ubiquitous throughout the Eukaryota (Vaucheret, 2006; Fire, 2007), suggesting that defense to viral infection via the processing of long dsRNA is well conserved. Key effector molecules involved in siRNA biogenesis do vary in both form and function and have been demonstrated to be targets of viral suppression in honeybees (De Smet et al., 2017). Both endo‐ and exo‐siRNAs are cytoplasmically processed by Dicer, a member of the RNAase III endonuclease family (Hammond et al., 2000; Bernstein et al., 2001). RNAase III enzymes are defined as having two RNAase III endonuclease domains and a helicase domain (Sontheimer, 2005). As well as having three conserved RNAase III motifs, the Dicers also contain a conserved RNA‐binding PAZ domain (Yan et al., 2003) and a DUF283 domain with unknown function (Dlakić, 2006).
Dicer's PAZ domain binds the 3′ overhangs of long cytoplasmic dsRNA molecules. The captured dsRNA is then brought into contact with Dicer's two RNAase III domains, each of which cleaves (or dices) a particular strand of the molecule. Dicing produces 21‐nt siRNA duplexes with short (2 nt) overhangs at the 3′ end on each strand (Elbashir et al., 2001a).
Many organisms (including humans) express a single isoform of Dicer (Zhang et al., 2002). Drosophila melanogaster expresses two Dicer variants (Dicer 1 and Dicer 2) that are reported to function in discrete gene silencing pathways (Lee et al., 2004; Tomari et al., 2007), though many details are as yet unclear. Dicer 2 binds and degrades long dsRNA destined to become siRNA, whereas Dicer 1 binds pre‐miRNA hairpin loops of ca. 60 nt and cleaves them, creating functional miRNAs duplexes. Dicer 2 is also instrumental for processing of siRNA in the small brown planthopper, Nilaparvata lugens (Stål) (Lan et al., 2016a) and the zigzag leafhopper, Recilia dorsalis Motschulsky (Lan et al., 2016b).
The RNA‐induced silencing complex (RISC)
RISCs are the functional components of all ncRNA‐mediated gene silencing pathways (Maniataki & Mourelatos, 2005; Rand et al., 2005; Hartig et al., 2007). RISCs can be defined as an Argonaute protein bound to a single strand of ncRNA. As there are several ncRNA and Argonaute species, the term RISC specifies a diverse group of ribonucleoprotein complexes.
Until complexed in a RISC, siRNAs have no effects upon gene expression. Formation of a RISC requires free siRNA to be captured by a RISC‐loading complex (RLC) and introduced to an Argonaute (Tomari et al., 2004; MacRae et al., 2008). After cleaving siRNA from long dsRNA, Dicer complexes with either transactivation response RNA‐binding protein (TRBP), or protein activator of PKR (PACT) to form the two human RLC variants (Haase et al., 2005; Lee et al., 2006, 2013; Lau et al., 2009). Variations in the 5′ terminal of TRBP and PACT may bias the binding affinities of human RLC variants toward either siRNA or miRNA, respectively (Lee et al., 2013). Dicer's participation in RISC loading is not required in all mammalian systems, as ΔDicer murine embryonic stem cells remain RLC competent (Murchison et al., 2005).
In Drosophila the canonical siRNA pathway utilizes an RLC formed by Dicer 2, R2D2 and TBP‐associated factor 11 (TAF11) (Liang et al., 2015). TAF11 is not necessary for RISC loading, as Dicer/R2D2 heterodimers form a competent RLC. Dicer and R2D2 bind opposite poles of siRNA before loading it into a RISC (Tomari et al., 2004). ∆R2D2 and ∆Dicer flies are therefore incapable of siRNA‐mediated gene silencing (Liu et al., 2006). Liang et al. (2015) suggest that an optimum RLC is formed when R2D2 and Dicer 2 form a tetrameric complex stabilized by TAF11. Tetrameric RLCs that include TAF11 display a 10‐fold increase in siRNA binding over Dicer/R2D2 heterodimers.
Dicer's role in siRNA biogenesis and RISC loading elicits potent gene silencing, in a manner which cannot yet be imitated precisely with artificially synthesized siRNA molecules. siRNAs that are enzymatically Diced from 240‐bp dsRNA constructs produce more effective gene silencing than artificially synthesized siRNA duplexes of equivalent sequence (Bolognesi et al., 2012). Whyard et al. (2009) also found that synthetic siRNA induced less potent RNAi than did enzymatically diced molecules.
siRNAs associate with Argonaute proteins from the Ago Clade
Based on analysis of nucleotide sequence homology, the Argonaute proteins form two clades. The Ago clade is the largest and took its name from Arabidopsis Ago1 mutants (Carmell et al., 2002). The Agos are bilobed proteins with a central PIWI endonuclease domain flanked by RNA‐binding PAZ (a feature shared with the Dicer enzymes), and MID domains at the N‐ and C‐terminals, respectively (Wang et al., 2008, 2009). A smaller Argonaute subclade (the PIWIs) was named for the Drosophila P‐element‐induced Wimpy testis protein (Aravin et al., 2006). Agos are expressed globally and associate with siRNA and miRNA to form RISCs. Until recently PIWIs (which associate with piRNAs) were thought to be restricted to the germline (Grishok et al., 2001; Morel et al., 2002; Tomari et al., 2007). However, evidence of their additional role in somatic gene silencing is now emerging (Morazzani et al., 2012; Schnettler et al., 2013).
Many organisms express a range of Ago proteins that associate with discrete ncRNA species. In D. melanogaster, for example, siRNA complexes with Ago2, miRNA associates with Ago1 (Tomari et al., 2007), and piRNA with PIWI proteins (Vagin et al., 2006; Malone et al., 2009). Although it is generally accepted that Ago2 is required for RNAi in the insects, silencing of Ago1 in Leptinotarsa decemlineata Say cells does inhibit gene silencing (Yoon et al., 2016). Humans, on the other hand, express four Ago proteins, all of which bind siRNA. However, only when complexed with Ago2 is siRNA capable of forming a functional RISC (Liu et al., 2004).
Guide strand genesis
Ago2 degrades a single ‘passenger’ strand of each siRNA duplex presented by an RLC (Matranga et al., 2005; Rand et al., 2005; Leuschner et al., 2006; Wang et al., 2009). In humans and D. melanogaster Ago2 initiates the release of the passenger strand by cleavage, creating two single‐stranded molecules of 9 and 12 nt (Matranga et al., 2005; Noland & Doudna, 2013). In humans the fragmented passenger strand is then degraded in the cytoplasm by C3PO (Ye et al., 2011). It also appears that C3PO aids passenger strand digestion in D. melanogaster and may also enhance gene silencing through RISC activation (Liu et al., 2009). Once the passenger strand has been released, the remaining ‘guide’ strand complexes with Ago2 to form the RISC.
The thermodynamic properties of duplexed siRNA molecules appear to influence which strand is destined to be integrated into a RISC. The two strands of the siRNA duplex have to be separated from each other by helicases, which try to unwind the duplex from both ends. The ends can have different stability depending on the GC content on the last 3–5 base pairs and the strand that has the 5′ end at the less strongly paired end has a higher chance to become the guide strand (Khvorova et al., 2003; Schwarz et al., 2003; Tomari et al., 2004). Both human RLC variants are capable of sensing the thermodynamics of duplexed siRNA and reorientating the molecule prior to RISC loading (Noland et al., 2011; Noland & Doudna, 2013), which may facilitate strand selection by Argonaute. The Dicer/R2D2 RLC seen in Drosophila also configures siRNA according the thermodynamics of the molecule (Liu et al., 2006).
RISCs utilize the guide strand to identify potential mRNA targets
Ago2's N‐terminal PAZ domain binds the 3′ end of the guide strand, whereas the C‐terminal MID domain binds the 5′ phosphate (Wang et al., 2008). The guide strand is orientated with its phosphate backbone toward Ago2's PIWI domain and the free nucleotides facing outwards. The RISC then utilizes the guide strand to scan cellular mRNA through Watson and Crick base pairing. Cognate mRNA, which base pairs with the guide, is targeted for knockdown (Filipowicz, 2005; Noland & Doudna, 2013). Each RISC is therefore capable of highly selective mRNA targeting based upon the nucleotide sequence of its intrinsic guide.
Cleavage of targeted mRNA
In the final stage of RNAi, cleavage of targeted mRNA occurs in the region bound by the center of the guide strand between residues 10 and 11 (Elbashir et al., 2001b; Haley & Zamore, 2004). The resulting 5′ and 3′ mRNA fragments are then degraded by discrete cytoplasmic enzymes (Orban & Izaurralde, 2005). Within Ago2's PIWI domain is an aspartate‐aspartate‐glutamate (DDE) motif which is conserved in RNAase‐H related enzymes (Song et al., 2004). This motif is critical for mRNA degradation, as mutation of these residues results in loss of slicing ability (Liu et al., 2004).
dsRNA as an experimental gene silencing device
In 1990, Napoli et al. (1990) developed petunias that expressed a hybrid chalcone synthase transgene (CHS). The authors predicted that expression of the transgene would supplement naturally occurring CHS and produce flowers with deep violet colouring (Napoli et al., 1990). Unexpectedly, 42% of the flowers exhibited an unpigmented, white phenotype. This led the authors to hypothesize that the transgene must somehow be inhibiting the expression of its naturally occurring orthologue.
Research into RNAi began following the work of Napoli et al. (1990). The first report of RNA being used to deliberately silence genes in an animal model came in 1995 when Guo & Kemphues (1995) injected C. elegans embryos with ssRNA designed to base pair with, and sequester, Par‐1 mRNA. Guo & Kemphues (1995) were successful in silencing Par‐1, but they were incorrect in their assumption that the underlying mechanism was triggered by ssRNA.
Using improved RNA preparation techniques, Fire et al. (1998) were able to show that Guo & Kemphues (1995) had contaminated their single‐stranded antisense RNAs with sense transcripts. Guo & Kemphues's (1995) ssRNAs had therefore base‐paired to form duplexes and entered the canonical siRNA pathway. Fire et al. (1998) were able to demonstrate that C. elegans when bathed in dsRNA silenced genes up to 100× more efficiently than when bathed in ssRNA. This experiment identified dsRNA as the critical effector molecules in previously described gene silencing experiments and was the first time dsRNA had purposefully been used to implement gene silencing. This finding was the starting point of all subsequent studies of RNAi.
Implementation of eRNAi in pest species
As outlined briefly above, microinjection of dsRNA would not be a viable method for treating the large numbers of insects required for SIT. However, it is thought that exposure to eRNAi might provide a suitable alternative. The susceptibility of target species to eRNAi is critically important and has been reviewed in depth by Baum & Roberts (2014). However, insects that are naturally recalcitrant to eRNAi are not necessarily outside consideration for this type of gene silencing as, although yet to be demonstrated in an insect model, methods such as electroporation can also be used to deliver dsRNA – e.g., as described in tick eggs (Karim et al., 2010; Ruiz et al., 2015), nymphs, and larvae (Lu et al., 2015). Various options are outlined below.
eRNAi delivery methods: larvae
To interrupt the sexual differentiation cascade in a manner that could be useful for SIT, RNAi must be implemented at the relevant critical developmental stages in eggs, embryos (Salvemini et al., 2009; Shukla & Palli, 2012; Liu et al., 2015), or early larvae (Whyard et al., 2015). Larvae are simple to target with eRNAi by ingestion as they eat steadily, volubly, and are generally less mobile than adults (hence can naturally take up dsRNA that is concentrated within local food sources). For aquatic larvae, dissolving dsRNA in solution and bathing the larvae within it, is the most common method of effecting gene silencing via eRNAi (Figueira‐Mansur et al., 2013; Singh et al., 2013; Whyard et al., 2015; Bona et al., 2016). dsRNA can be delivered to non‐aquatic larvae: (1) topically via droplet feeding (Toprak et al., 2013), (2) by inducing the larvae to feed upon dsRNA‐expressing transgenic plants (Xiong et al., 2013; Mamta et al., 2015; Tian et al., 2015; Hu et al., 2016), (3) by feeding larvae dsRNA‐expressing transgenic bacteria (Zhu et al., 2011; Yang & Han, 2014; Li et al., 2015c), and (4) by feeding larvae naked dsRNA overlaid onto an artificial diet (Asokan et al., 2014; Yang & Han, 2014; Hu et al., 2016). Non‐aquatic larvae, or those that develop in relatively anoxic conditions, can also be bathed in dsRNA solution, but the timing of exposure is critical to avoid drowning (Whyard et al., 2009). Choi et al. (2012) also report delivery of dsRNA via parental feeding in a study in which nurse ant workers were fed with dsRNA that was then passed to larvae via regurgitation.
eRNAi delivery methods: adults
Genes that can induce sterility when knocked down can be targeted in adult insects for use with SIT. eRNAi has been demonstrated to successfully achieve gene silencing in adults following ingestion of: (1) dsRNA‐expressing transgenic plants (Coleman et al., 2015; Tzin et al., 2015; Malik et al., 2016), (2) dsRNA‐expressing transgenic bacteria (Li et al., 2011; Taracena et al., 2015; Whitten et al., 2015), (3) dsRNA dissolved in solution (Coy et al., 2012; Ratzka et al., 2013; Shim et al., 2015), and (4) naked dsRNA overlaid on diet (Yi et al., 2014; Zheng et al., 2015). In addition, topical application to adults of dsRNA (Pridgeon et al., 2008; Killiny et al., 2014; Amiri et al., 2015) and infection with transgenic fungi (Chen et al., 2015) are reported. All methods have the potential for use in SIT development. However, the use of transgenic plants may be limited by the feeding habits of target pests, and fungi also need to be tested for their potential to infect unintended secondary targets.
An important consideration for eRNAi silencing for insect control is the feasibility of producing and delivering the required amount of dsRNA. Both in vitro and in vivo methods for producing dsRNA for insect control have been tested, as described below.
In vitro dsRNA synthesis
The T7 RNA polymerase (from the T7 bacteriophage) is a highly selective enzyme that enables rapid synthesis of RNA sequences (Tabor, 2001). For in vitro production of dsRNA, linear DNA sequences that code for both sense and antisense RNA transcripts flanked by the 20‐nt T7 promoter are transcribed by incubation with T7 polymerase (Singh et al., 2013; Liu et al., 2015; Shim et al., 2015; Whyard et al., 2015). Cognate ssRNA transcripts then base pair to form dsRNA that can be used for eRNAi experiments.
In vivo dsRNA synthesis by bacteria
dsRNA can be synthesized in vivo by bacteria themselves using transgenic HT115 Escherichia coli (Migula) Castellani & Chalmers (Kamath et al., 2001). The HT115 genome has been modified to be RNase deficient and to contain a T7 polymerase under the control of lactose regulatory elements. Generally, target sequences flanked by two T7 promoters at each side are introduced to L4440 plasmid vectors by ligation. The plasmid is then transformed into HT115 bacteria and target DNA sequences are transcribed by T7 polymerases induced by the allolactose mimic IPTG (Whyard et al., 2009, 2015; Zhu et al., 2011; Yang & Han, 2014; Taracena et al., 2015). A limitation of this method is that, once introduced to target insects, the effect is transient as the HT115 bacteria fail to colonize the gut and become established in the insect gut microbiome.
Modified symbiotic bacteria have recently been utilized as an alternative to HT115 E. coli (Whitten et al., 2015). In this study, microbes from the microbiome of target species were reprogrammed to have similar properties to HT115 (in that they were RNAse‐deficient), but RNA sequences were constitutively active rather than being inducible. These symbiotic bacteria were able to repopulate the gut of target insects and induce a long term silencing effect. These results suggest that there is great potential to genetically engineer naturally occurring bacteria in the gut microbiomes of pest species for control purposes.
In vivo dsRNA synthesis by plants
The nuclear genome of plants can be modified using Agrobacterium tumefaciens Smith & Townsend (De Block et al., 1984; Horsch et al., 1984) to express non‐endogenous dsRNA. dsRNA constructs can be expressed as either a single sequence which forms a long hairpin (hpRNA) (Xiong et al., 2013; Guo et al., 2014; Mamta et al., 2015), or two separate complementary transcripts which base pair in the cytoplasm (Kumar et al., 2012). However, transfer of target sequences into the genome of plant hosts is unpredictable and dsRNA abundance in similarly prepared transgenic plants can vary by up to 900% (Tian et al., 2015).
The genome of plant chloroplasts can also be programmed to synthesize non‐endogenous dsRNA (Jin et al., 2015; Zhang et al., 2015a). Due to the prodigious metabolic output of these organelles they are capable of rapid production of large amounts of effector dsRNA molecules. As for the gut microbiota, there is potential for engineering chloroplasts in this manner for application to insects through eRNAi. In the next sections we consider the design features of dsRNAs that may render them useful for control.
Optimum dsRNA construct design: length and GC content
There is a minimum length threshold (MLT) at which dsRNA can induce eRNAi. The MLT has been demonstrated to be ca. 60 bp in several insects (Bolognesi et al., 2012; Miller et al., 2012; Ivashuta et al., 2015), although Miyata et al. (2014) reported an MLT of ca. 100 bp. The MLT for eRNAi is defined by the minimum length of dsRNA that can be absorbed by the intestine. However, distal tissues may be capable of absorbing shorter transcripts. Ivashuta et al. (2015) report an MLT of 60 bp in Diabrotica virgifera virgifera LeConte, and the uptake of a 21‐bp siRNAs by the fat body of this insect.
Once the MLT has been met, dsRNA construct length is not an accurate predictor of RNAi potency, as constructs of similar length can elicit diverse silencing effects (Toprak et al., 2013; Asokan et al., 2014). Most RNAi research in insects is carried out using dsRNA constructs of between 200–500 bp (Table 1), although successful silencing has been achieved using constructs of up to 1 800 bp (Baum et al., 2007).
Table 1.
A selection of examples of the effectiveness of eRNAi across diverse insect taxa. The studies included highlight variation in the strength of silencing that can be induced via different methods of dsRNA delivery in various taxa and the differences observed in temporal effects
| Species | dsRNA delivery method | Construct length/bp | Genes targeted | Temporal effects of RNAi | Quantification of gene knockdown | Reference | |
|---|---|---|---|---|---|---|---|
| Hymenoptera | Camponotus floridanus (Buckley) | Adults fed dsRNA dissolved in sucrose solution for up to 15 days | ≈ 400 | Adults: peptidoglycan recognition protein (PGRP‐LB) | Maximum gene silencing after 5 days | ≈ 100% | Ratzka et al. (2013) |
| Solenopsis invicta Buren | dsRNA fed to larvae via nurse workers for 12 days | 496 | Pheromone biosynthesis activating neuropeptide (PBAN) | After 21 days mortality increased by 50% | Not known | Choi et al. (2012) | |
| Coleoptera | Diabrotica virgifera virgifera | Embryos targeted via parental RNAi; adults fed dsRNA overlaid on artificial diet for 10 days | 352, 405 | Brahma (bhm) and Hunchback (hb) | 10 days after egg laying hatching rates were 0% (bhm) and 2.4% (hb) | 99% silencing of brahma in eggs; 85.3% silencing of hunchback in eggs | Khajuria et al. (2015) |
| Leptinotarsa decemlineata | L2 larvae fed dsRNA or transgenic bacteria overlaid on potato leaves | 200–400 | 5× Housekeeping genes | Bacteria‐treated larvae exhibit higher mortality than naked dsRNA treatment, 12 days after feeding | Bacteria‐treated: 59–91%; dsRNA: 61–93% (both gene‐dependent) | Zhu et al. (2011) | |
| D. v. virgifera | L1 larvae fed dsRNA overlaid on diet for up to 8 days | 250, 500, 750, 1000 | Ebony and laccase 2 (lac2) | ≈ 90% silencing after 2 days | Lac2 ≈ 100%; ebony ≈ 95% | Miyata et al. (2014) | |
| Hemiptera | Myzus persicae (Sulzer) | Adults fed on transgenic plants for up to16 days | Not known | Receptor of Activated Kinase C (Rack1), MpPIntO2 & MpC002 | Maximum effect seen after 8 days. Treated insects demonstrate gene silencing for up to 6 days post‐feeding. Progeny of treated insects demonstrate gene silencing for up to10 days post‐feeding | ≈ 70% for all genes | Coleman et al. (2015) |
| M. persicae/Bactericera cockerelli (Šulc) | Adults fed on transgenic plants for up to 8 days | 250–500 | Aquaporin (AQP), sucrase (SUC), and sugar transporter (ST4) | B. cockerelli: osmotic pressure of haemolymph varied significantly for 8 days; M. persicae: gene expression altered for 7 days | B. cockerelli: no significant silencing of any genes; M. persicae: significant silencing of all genes | Tzin et al. (2015) | |
| Rhodnius prolixus Stål | Adult females fed ≈ 154.2 ng of dsRNA incorporated in transgenic bacteria | 375, 453 | Rhodnius heme‐binding protein (RHBP) and catalase (CAT) | RHBP: peak silencing after 3 days of feeding, no effect 10 days after feeding; CAT: peak silencing after 5 days of feeding | RHBP: 99.6%; CAT: 96% silencing | Taracena et al. (2015) | |
| Diaphorina citri Kuwayama | Topical application of dsRNA dissolved in solution to adults | Not known | 5× Cytochrome p450 genes | Significant lack of protein 8 days after treatment | 50–100% silencing (gene dependent) | Killiny et al. (2014) | |
| Bemisia tabaci (Gennadius) | Adults fed dsRNA dissolved in sucrose solution for 1 day. | 279 | Heat shock protein 70 (hsp70) | Gene silencing for up to 3 days after feeding. | 100% silencing | Shim et al. (2015) | |
| Lepidoptera | Mamestra configurata Walker | dsRNA applied topically to L1 or L4 larvae, which are then fed with leaf discs overlaid with dsRNA for up to 2 days | 500 | Chitin deacetylase 1 (CDA1) | Larvae silence CDA1 1 (L1) and 1.5 (L4) days after treatment | L1: ≈100% silencing; L4: silencing not quantified | Toprak et al. (2013) |
| Helicoverpa armigera | Larvae fed transgenic plants or bacteria for 7 days | 400–600 | Larvae: molt regulating transcription factor (HR3) | 2‐day latency of effect; larvae fed on transgenic plants have 70% less mass than controls after 6 days | Bacteria: ≈ 80%; plants: ≈ 85% | Xiong et al. (2013) | |
| H. armigera | Larvae fed (all instars) transgenic bacteria and dsRNA overlaid on artificial diet | 562, 450 | Ultraspiracle protein (Ups) and ecdysone receptor gene (EcR) | Surviving larvae were assayed by qRT‐PCR 5 days after treatment | 60% silencing of USP via continuous bacterial feeding | Yang & Han (2014) | |
| Diptera | Aedes aegypti | Topical application of dsRNA to adult females | 252, 436, 556 | 3× Inhibitor of apoptosis (IAP) genes | Mosquitos assessed 1 day after treatment | 33–87.5% silencing (gene dependent) | Pridgeon et al. (2008) |
| Anopheles gambiae Giles | L3 larvae fed dsRNA complexed in chitosan nanoparticles | Not known | Chitin synthase 1 (CHS1) and chitin synthase 2 (CHS2) | After 4 days of treatment with CHS1 (but not CHS2), larvae silenced both intended target genes | CHS1: 62.8% silencing of CHS1 and 57.9% of CHS2; CHS2: 63.4% | Zhang et al. (2010) | |
| Bactrocera dorsalis | Adults fed dsRNA overlaid on artificial diet until death | Not known | Sex peptide receptor (spr) | Mean life span reduced by 26 days in treated flies | 52% | Zheng et al. (2015) | |
| B. dorsalis | Adults fed dsRNA overlaid on artificial diet for up to 14 days | 297, 394 | Odorant receptor (Bdor) and odorant coreceptor (Orco); targets silenced individually and simultaneously | 4‐day latency of effect, peak effect after 7 days | 70% when both targets applied simultaneously | Yi et al. (2014) | |
| Drosophila melanogaster | L1 larvae soaked in PBS buffer containing dsRNA bound in liposomal vectors for 1 h | ≈ 400 | β‐Glucuronidase gene (gus) | Flies assessed 1 day after feeding | Lipofamectine 2000 vectors: 53% | Whyard et al. (2009) |
The GC content of dsRNA negatively correlates with eRNAi efficiency (Reynolds et al., 2004; Chan et al., 2009). GC bonds are more stable than AU bonds and less prone to unwinding by Dicer's helicase domain.
Optimum dsRNA construct design: target sequence
Specific nucleotide sequences within targeted mRNAs are intrinsically susceptible to RISC cleavage. Bolognesi et al. (2012) report that a single homologous 21‐bp sequence can silence mRNA as efficiently as a 60‐bp construct of 100% homology. RNAi potency is therefore governed partly by the quality of 21‐nt sequences contained within dsRNA constructs.
Discrete dsRNA constructs targeting different regions of the same mRNA molecule have been demonstrated to elicit variable silencing effects (Xiong et al., 2013; Tian et al., 2015). Neither the pole nor the mid‐region of mRNA appears to be intrinsically prone to cleavage, as susceptibility has been reported at various regions of targeted molecules (Mao & Zeng, 2012; Xiong et al., 2013). Asokan et al. (2014) targeted five unrelated mRNAs in Helicoverpa armigera (Hübner) and observed variation in silencing efficiency from 21 to 95%.
Sfold software (Ding et al., 2005) is reported to predict the regional susceptibility of mRNA to RISC cleavage prior to construct design, though data from Xiong et al. (2013) showed predictive success to be variable. Regional susceptibility to cleavage is likely to be defined largely by the secondary structure of the target molecule. RNAi potency is inversely proportional to the amount of hydrogen bonds formed between target and non‐target sequences (Luo & Chang, 2004). This implies that targets that are tightly bonded to local sequences are unavailable to the RISC complex, rendering gene silencing inefficient.
Optimum dsRNA construct design: off‐target effects
Non‐target mRNA with precise homology to the guide strand of a RISC will inevitably be targeted and degraded. Hence, it is important to consider the potential for inadvertently silencing any other genes within the same or different species that contain these sequences. Such off‐target effects (OTEs) can affect endogenous genes of the experimental target (Kulkarni et al., 2006; Ma et al., 2006; Zhang et al., 2010; Toprak et al., 2013), predators of the target species following consumption (Garbian et al., 2012), and closely related species (Zhu et al., 2012). Singh et al. (2013) found that sequences of continuous homology greater than 19 nt were required to induce OTEs in target (Aedes) and non‐target (Drosophila) species. Ulrich et al. (2015) report that OTEs are equally probable in conserved and non‐conserved amino acid sequences.
It is important to minimize the potential for OTEs by careful design of dsRNAs. ‘E‐RNAi’ (Horn & Boutros, 2010) and ‘SnapDragon’ (Harvard Medical School, 2016) are examples of software that automatically design dsRNAs for use with RNAi and search for OTEs in a selection of well‐referenced genomes. If dsRNAs are designed manually then dsCheck (Naito et al., 2005) can be used to predict potential OTEs. However, it is difficult to fully anticipate off‐target matching for sequences that have not yet been described.
Genetic attributes which facilitate eRNAi – the Caenorhabditis elegans example
The nematode Caenorhabditis elegans (Maupas) was the first species in which RNAi was successfully implemented (Fire et al., 1998) and has been used extensively to investigate genetic mechanisms underlying gene silencing. This research has uncovered five systemic interference defective (SID) genes that facilitate RNAi (Feinberg & Hunter, 2003; Jose & Hunter, 2007; Jose et al., 2012). SID2 is an intestinal transmembrane protein that is thought to independently endocytose vesicular dsRNA from the gut lumen, before it is processed in the cytoplasm by Dicer (McEwan et al., 2012). SID2 proteins are essential for eRNAi in C. elegans as they allow for passage of ingested dsRNA molecules and enable eRNAi when transgenically expressed in recalcitrant species (Winston et al., 2007). SID1 is a ubiquitously expressed transmembrane protein that is essential for systemic RNAi (Winston et al., 2002; Jose et al., 2009). SID1's precise mode of action is yet to be elucidated, but it transmits silencing signals between cells, either in the form of long dsRNAs, free siRNAs, or siRNA bound by RISCs.
In C. elegans, RNAi is propagated by RNA‐dependent RNA polymerases (RdRp) (Sijen et al., 2001). RdRps bind primary cytoplasmic RNA transcripts and utilize them to synthesize secondary dsRNAs, which then re‐enter the siRNA pathway extending the period of gene knockdown (Pak & Fire, 2007). Many viruses also encode RdRps, which allow for the proliferation of viral RNAs (Pan et al., 2016).
The wealth of information about the mechanism of RNAi gained from the study of C. elegans has been of great use for elucidating homologous mechanisms in pest species. The transfer of enabling mechanisms, such as SID1 gene functionality, to species that lack them has also provided insights into gene silencing mechanisms that might exist in some pest species.
Genetic attributes that facilitate eRNAi in the insects
Orthologues of SID genes are found across insect taxa but are absent from the Diptera (Huvenne & Smagghe, 2010), members of which represent some of the world's most significant agricultural and medical pests. An orthologue of SID1 facilitates systemic RNAi in Apis mellifera L. (Aronstein et al., 2006) and Miyata et al. (2014) suggest that two SID orthologues are involved in, if not essential to, eRNAi in D. v. virgifera. On this basis, the presence or absence of SIDs has been used by some researchers to predict the capacity of different insect species to effect gene silencing via eRNAi (Tomoyasu et al., 2008; Tian et al., 2009). However, the presence of SIDs is not fully predictive of eRNAi potential – several studies have demonstrated that dipterans, which lack SIDs as noted above, are eRNAi competent (Table 1). In contrast, Bombyx mori (L.) possesses three SID orthologues, yet is not competent for eRNAi (Li et al., 2015d).
SIDs are not the only molecules that contribute to dsRNA uptake as eRNAi is facilitated by endocytic pathways in some insect species. For example, Bactrocera dorsalis (Hendel) (Li et al., 2015b) and Tribolium castaneum (Herbst) (Xiao et al., 2015) are refractory to eRNAi following challenge with the endocytic inhibitor Bafilomycin A1 (Xu et al., 2003). Refractoriness to eRNAi can also be induced in Bactrocera and Tribolium if orthologues of the chc (clathrin heavy chain) gene (Bazinet et al., 1993) are downregulated (Li et al., 2015b; Xiao et al., 2015). It has recently been suggested that SIDs and endocytic mediators synergistically facilitate eRNAi in L. decemlineata (Cappelle et al., 2016). Scavenger receptors have also been demonstrated to facilitate systemic RNAi in D. melanogaster (Saleh et al., 2009) and eRNAi in Schistocerca gregaria Forsskål (Wynant et al., 2014).
Recent evidence suggests that an intracellular mode of dsRNA degradation, other than the siRNA pathway, may exist in some insect species (Shukla et al., 2016). Shukla et al. (2016) demonstrated that cell lines of L. decemlineata and Spodoptera frugiperda (JE Smith) were both capable of dsRNA uptake, although only the cells of Leptinotarsa produced 21‐bp siRNA‐like transcripts. Apparently dsRNA was degraded in endosomes within the cells of Spodoptera, as demonstrated by pH‐induced fluorescence of CypHer5E‐labelled molecules. Accordingly, Yoon et al. (2016) report that dsRNA may escape endosomes through acidification in L. decemlineata, facilitating induction of the RNAi process in that insect. These types of practical studies, along with comparative transcriptomic analyses (such as Swevers et al., 2013), may help us to understand the divergence in eRNAi potency between insects.
Orthologues of RdRps have not been reported in the Insecta. Several insects can nevertheless exhibit sustained RNAi for prolonged periods (Paim et al., 2013; Coleman et al., 2015; Khajuria et al., 2015), suggesting a system of signal amplification. Hemipterans appear to have a robust RNAi amplification system, as gene silencing has been demonstrated for 4 days (Rebijith et al., 2016), 6 days (Coleman et al., 2015), and 8 days (Tzin et al., 2015) after feeding with dsRNA. Coleman et al. (2015) also report that nymphs born from RNA‐treated mothers exhibited RNAi for 10 days post‐feeding, suggesting that genetic variation between life‐history stages might influence signal amplification.
Insects that produce RNAses in salivary and midgut secretions or in haemolymph can degrade dsRNAs and thus limit the extent of gene silencing (Allen & Walker, 2012; Liu et al., 2012; Garbutt et al., 2013; Yang & Han, 2014; Shukla et al., 2016). Yang & Han (2014) suggest that RNAi is more efficient in H. armigera if dsRNAs are encapsulated in bacteria when they traverse the gut, as this bypasses potent digestive RNAases in midgut secretions. Das et al. (2015) present a similar idea, but suggest that vectoring dsRNA in carbon quantum dot nanoparticles provides protection, not from nucleolytic enzymes, but from damage incurred through extreme pH in the alimentary canal of A. aegypti.
Conclusion
In this review we have described the detailed mechanisms underlying gene silencing by dsRNA, and considered the use of this approach for use with SIT. Currently, the available data are scant and insufficient to design all aspects of eRNAi studies in pest species in a predictive context. We have emphasized several factors that must be considered in the design and implementation of such techniques (Table 2) in order to try to address these omissions.
Knowledge of eRNAi in key areas, such as the most basic mechanisms that enable insects to acquire dsRNA from their environment, is lacking. SIDs and endocytosis both play roles individually and synergistically, but overall the picture of their modes of action is far from clear. Packaging of dsRNA in intermediate carriers such as bacteria or nanoparticles may overcome refractoriness to eRNAi in some cases. However, certain insects may remain refractory to eRNAi even if dsRNA is successfully packaged and transported across the gut, as discrete modes of dsRNA degradation such as endosomal acquisition may mitigate the silencing process.
SIT strategies rely on mass rearing, which in some cases has knock‐on effects for insect quality and performance (Sørensen et al., 2012). The consequences of mass rearing manifest differently across taxa, which is why Chambers (1977) suggested comprehensive quality control measures for all such programs. SIT individuals generated (by any means) from mass‐reared populations are likely to perform differently than untreated controls. All SIT techniques should be judged according to the performance of individuals in the field, which very often will differ to those in the laboratory or factory (Mayer et al., 1998; Carvalho et al., 2015). The goal of eRNAi‐SIT is to produce mass‐reared insects that are at least equal in quality to the currently available alternatives.
The potential of an eRNAi approach is being increasingly realized and groups such as the SITplus partnership are developing this technology to combat the spread of pest insects (CSIRO, 2015). The production of dsRNA for large‐scale eRNAi treatments may be expensive. HT115 E. coli (see ‘In vivo dsRNA synthesis’ section above) may represent the most viable option currently available, but economy of scale does present a challenge that needs to be addressed.
As suggested by the transformer‐2/innexin‐5 model, eRNAi could be used in the future to successfully implement gene silencing, and create insects for application of SIT in the field. There is evidence that other gene targets could also be utilized in an eRNAi‐SIT system. A possible outcome when the sexual differentiation cascade is targeted with RNAi is a combination of arrested phenotypic female development in some individuals, and sex‐reversal in others. This has been demonstrated by Shukla & Palli (2012) in T. castaneum, where parental RNAi of transformer produced a cohort of 91.1% males, 8.9% pseudomales, and 0% females. This outcome is not optimal for SIT as nearly half the population was lost, but it did produce a compatible male‐only cohort. More recently, eRNAi of spermatogenic targets has been demonstrated to induce sterility by up to 60% in B. dorsalis while maintaining mating competitiveness (Dong et al., 2016).
Acknowledgements
We thank the BBSRC for funding (BB/N50418X/1, Industrial Case Partnership PhD studentship to Oxitec and TC). NIM is an employee of Oxitec, a company developing genetics‐based control methods against insects of agricultural and medical significance.
References
- Abd El Halim HM, Alshukri BMH, Ahmad MS, Nakasu EYT, Awwad MH et al. (2016) RNAi‐mediated knockdown of the voltage gated sodium ion channel TcNav causes mortality in Tribolium castaneum . Scientific Reports 6: 29301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen ML & Walker WB (2012) Saliva of Lygus lineolaris digests double stranded ribonucleic acids. Journal of Insect Physiology 58: 391–396. [DOI] [PubMed] [Google Scholar]
- Alphey L, Benedict M, Bellini R, Clark GG, Dame DA et al. (2010) Sterile‐insect methods for control of mosquito‐borne diseases: an analysis. Vector Borne and Zoonotic Diseases 10: 295–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri A, Bandani AR & Alizadeh H (2015) Molecular identification of cysteine and trypsin protease, effect of different hosts on protease expression, and rnai mediated silencing of cysteine protease gene in the sunn pest. Archives of Insect Biochemistry and Physiology 91: 189–209. [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos‐Quintana M, Landgraf P et al. (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207. [DOI] [PubMed] [Google Scholar]
- Aronstein K, Pankiw T & Saldivar E (2006) SID‐I is implicated in systemic gene silencing in the honey bee. Journal of Apicultural Research 45: 20–24. [Google Scholar]
- Asokan R, Sharath Chandra G, Manamohan M, Krishna Kumar NK & Sita T (2014) Response of various target genes to diet‐delivered dsRNA mediated RNA interference in the cotton bollworm, Helicoverpa armigera . Journal of Pest Science 87: 163–172. [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP & Blelloch R (2008) Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor‐independent, dicer‐dependent small RNAs. Genes and Development 22: 2773–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Baum JA & Roberts JK (2014) Progress towards RNAi‐mediated insect pest management Advances in Insect Physiology, Vol. 47 (eds. by Tarlochan SD. & Sarjeet SG.), pp. 249–295. Academic Press, London, UK. [Google Scholar]
- Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P et al. (2007) Control of coleopteran insect pests through RNA interference. Nature Biotechnology 25: 1322–1326. [DOI] [PubMed] [Google Scholar]
- Bazinet C, Katzen AL, Morgan M, Mahowald AP & Lemmon SK (1993) The Drosophila clathrin heavy chain gene clathrin function is essential in a multicellular organism. Genetics 134: 1119–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM & Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Bolognesi R, Ramaseshadri P, Anderson J, Bachman P, Clinton W et al. (2012) Characterizing the mechanism of action of double‐stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS ONE 7: e47534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bona AC, Chitolina RF, Fermino ML, de Castro Poncio L, Weiss A et al. (2016) Larval application of sodium channel homologous dsRNA restores pyrethroid insecticide susceptibility in a resistant adult mosquito population. Parasites and Vectors 9: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briceño RD & Eberhard WG (2003) Decisions during courtship by male and female medflies (Diptera, Tephritidae): correlated changes in male behavior and female acceptance criteria in mass‐reared flies. Florida Entomologist 85: 14–31. [Google Scholar]
- Cao J, Liu Y, Yang Y, Zhang H, Li Z et al. (2015) Molecular characterization and functional analysis of the ultraspiracle (USP) in the oriental fruit moth Grapholita molesta (Lepidoptera: Olethreutidae). Comparative Biochemistry and Physiology B 190: 54–62. [DOI] [PubMed] [Google Scholar]
- Cappelle K, de Oliveira CF, Van Eynde B, Christiaens O & Smagghe G (2016) The involvement of clathrin‐mediated endocytosis and two Sid‐1‐like transmembrane proteins in double‐stranded RNA uptake in the Colorado potato beetle midgut. Insect Molecular Biology 25: 315–323. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ & Hannon GJ (2002) The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes and Development 16: 2733–2742. [DOI] [PubMed] [Google Scholar]
- Carvalho DO, McKemey AR, Garziera L, Lacroix R, Donnelly CA et al. (2015) Suppression of a field population of Aedes aegypti in Brazil by sustained release of transgenic male mosquitoes. PLoS Neglected Tropical Diseases 9: e0003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers DL (1977) Quality control in mass rearing. Annual Review of Entomology 22: 289–308. [Google Scholar]
- Chan CY, Carmack CS, Long DD, Maliyekkel A, Shao Y et al. (2009) A structural interpretation of the effect of GC‐content on efficiency of RNA interference. BMC Bioinformatics 10(Suppl 1): S33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li L, Hu Q, Zhang B, Wu W et al. (2015) Expression of dsRNA in recombinant Isaria fumosorosea strain targets the TLR7 gene in Bemisia tabaci . BMC Biotechnology 15: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi MY, Vander Meer RK, Coy M & Scharf ME (2012) Phenotypic impacts of PBAN RNA interference in an ant, Solenopsis invicta, and a moth, Helicoverpa zea . Journal of Insect Physiology 58: 1159–1165. [DOI] [PubMed] [Google Scholar]
- Christiaens O, Prentice K, Pertry I, Ghislain M, Bailey A et al. (2016) RNA interference: a promising biopesticide strategy against the African sweetpotato weevil Cylas brunneus . Scientific Reports 6: 38836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman AD, Wouters RHM, Mugford ST & Hogenhout SA (2015) Persistence and transgenerational effect of plant‐mediated RNAi in aphids. Journal of Experimental Botany 66: 541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy MR, Sanscrainte ND, Chalaire KC, Inberg A, Maayan I et al. (2012) Gene silencing in adult Aedes aegypti mosquitoes through oral delivery of double‐stranded RNA. Journal of Applied Entomology 136: 741–748. [Google Scholar]
- CSIRO (2015) Partnering to find a solution to Australia's Qfly problem. Available at: http://www.csiro.au/en/Research/BF/Areas/Protecting-Australias-agricultural-industries/Robot-technology/SITplus (accessed 06 October 2016).
- Das S, Debnath N, Cui Y, Unrine J & Palli SR (2015) Chitosan, carbon quantum dot, and silica nanoparticle mediated dsRNA delivery for gene silencing in Aedes aegypti: a comparative analysis. ACS Applied Materials and Interfaces 7: 19530–19535. [DOI] [PubMed] [Google Scholar]
- De Block M, Herrera‐Estrella L, Van Montagu M, Schell J & Zambryski P (1984) Expression of foreign genes in regenerated plants and in their progeny. EMBO Journal 3: 1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet L, Ravoet J, Wenseleers T & de Graaf DC (2017) Expression of key components of the RNAi machinery are suppressed in Apis mellifera that suffer a high virus infection. Entomological Science 20: 76–85. [Google Scholar]
- Ding Y, Chi YC & Lawrence CE (2005) RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA 11: 1157–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakić M (2006) DUF283 domain of Dicer proteins has a double‐stranded RNA‐binding fold. Bioinformatics 22: 2711–2714. [DOI] [PubMed] [Google Scholar]
- Dong YC, Wang ZJ, Chen ZZ, Clarke AR & Niu CY (2016) Bactrocera dorsalis male sterilization by targeted RNA interference of spermatogenesis: empowering sterile insect technique programs. Scientific Reports 6: 35750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K & Tuschl T (2001a) Duplexes of 21‐nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W & Tuschl T (2001b) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO Journal 20: 6877–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg EH & Hunter CP (2003) Transport of dsRNA into cells by the transmembrane protein SID‐1. Science 301: 1545–1547. [DOI] [PubMed] [Google Scholar]
- Figueira‐Mansur J, Ferreira‐Pereira A, Mansur JF, Franco TA, Alvarenga ESL et al. (2013) Silencing of P‐glycoprotein increases mortality in temephos‐treated Aedes aegypti larvae. Insect Molecular Biology 22: 648–658. [DOI] [PubMed] [Google Scholar]
- Filipowicz W (2005) RNAi: the nuts and bolts of the RISC machine. Cell 122: 17–20. [DOI] [PubMed] [Google Scholar]
- Fire AZ (2007) Gene silencing by double‐stranded RNA. Cell Death and Differentiation 14: 1998–2012. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu SQ, Montgomery MK, Kostas SA, Driver SE & Mello CC (1998) Potent and specific genetic interference by double‐stranded RNA in Caenorhabditis elegans . Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Gabrieli P, Scolari F, Di Cosimo A, Savini G, Fumagalli M et al. (2016) Sperm‐less males modulate female behaviour in Ceratitis capitata (Diptera: Tephritidae). Insect Biochemistry and Molecular Biology 79: 13–26. [DOI] [PubMed] [Google Scholar]
- Garbian Y, Maori E, Kalev H, Shafir S & Sela I (2012) Bidirectional transfer of RNAi between honey bee and Varroa destructor: Varroa gene silencing reduces Varroa population. PLoS Pathogens 8: e1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbutt JS, Bellés X, Richards EH & Reynolds SE (2013) Persistence of double‐stranded RNA in insect hemolymph as a potential determiner of RNA interference success: evidence from Manduca sexta and Blattella germanica . Journal of Insect Physiology 59: 171–178. [DOI] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP & Vaucheret H (2005) Partially redundant functions of arabidopsis DICER‐like enzymes and a role for DCL4 in producing trans‐Acting siRNAs. Current Biology 15: 1494–1500. [DOI] [PubMed] [Google Scholar]
- Gorman K, Young J, Pineda L, Marquez R, Sosa N et al. (2016) Short‐term suppression of Aedes aegypti using genetic control does not facilitate Aedes albopictus . Pest Management Science 72: 618–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S et al. (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- Gross M (2013) EU ban puts spotlight on complex effects of neonicotinoids. Current Biology 23: R462–R464. [DOI] [PubMed] [Google Scholar]
- Gu L & Knipple DC (2013) Recent advances in RNA interference research in insects: implications for future insect pest management strategies. Crop Protection 45: 36–40. [Google Scholar]
- Guerfali MM, Parker A, Fadhl S, Hemdane H, Raies A & Chevrier C (2011) Fitness and reproductive potential of irradiated mass‐reared mediterranean fruit fly males Ceratitis capitata (Diptera: Tephritidae): lowering radiation doses. Florida Entomologist 94: 1042–1050. [Google Scholar]
- Guo S & Kemphues KJ (1995) par‐1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell 81: 611–620. [DOI] [PubMed] [Google Scholar]
- Guo H, Song X, Wang G, Yang K, Wang Y et al. (2014) Plant‐generated artificial small RNAs mediated aphid resistance. PLoS ONE 9: e97410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kang S, Zhu X, Xia J, Wu Q et al. (2015) The novel ABC transporter ABCH1 is a potential target for RNAi‐based insect pest control and resistance management. Scientific Reports 5: 13728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang HD, Laine S, Sack R et al. (2005) TRBP, a regulator of cellular PKR and HIV‐1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Reports 6: 961–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B & Zamore PD (2004) Kinetic analysis of the RNAi enzyme complex. Nature Structural and Molecular Biology 11: 599–606. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D & Hannon GJ (2000) An RNA‐directed nuclease mediates post‐transcriptional gene silencing in Drosophila cells. Nature 404: 293–296. [DOI] [PubMed] [Google Scholar]
- Hannon GJ (2002) RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Harris AF, Nimmo D, McKemey AR, Kelly N, Scaife S et al. (2011) Field performance of engineered male mosquitoes. Nature Biotechnology 29: 1034–1037. [DOI] [PubMed] [Google Scholar]
- Hartig JV, Tomari Y & Foerstemann K (2007) piRNAs ‐ the ancient hunters of genome invaders. Genes and Development 21: 1707–1713. [DOI] [PubMed] [Google Scholar]
- Harvard Medical School (2016) SnapDragon. Available at: www.flyrnai.org (accessed 08 August 2016).
- Hooper GHS (1972) Sterilization of the Mediterranean fruit fly with gamma radiation: effect on male competitiveness and change in fertility of females alternately mated with irradiated and untreated males. Journal of Economic Entomology 65: 1–6. [DOI] [PubMed] [Google Scholar]
- Horn T & Boutros M (2010) E‐RNAi: a web application for the multi‐species design of RNAi reagents‐2010 update. Nucleic Acids Research 38: W332–W339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch RB, Fraley RT, Rogers SG, Sanders PR, Lloyd A & Hoffmann N (1984) Inheritance of functional foreign genes in plants. Science 223: 496–498. [DOI] [PubMed] [Google Scholar]
- Hu J, Liu J & Corey DR (2010) Allele‐selective inhibition of huntingtin expression by switching to an miRNA‐like RNAi mechanism. Chemistry and Biology 17: 1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Richtman NM, Zhao JZ, Duncan KE, Niu X et al. (2016) Discovery of midgut genes for the RNA interference control of corn rootworm. Scientific Reports 6: 30542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huvenne H & Smagghe G (2010) Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: a review. Journal of Insect Physiology 56: 227–235. [DOI] [PubMed] [Google Scholar]
- Ivashuta S, Zhang Y, Wiggins BE, Ramaseshadri P, Segers GC et al. (2015) Environmental RNAi in herbivorous insects. RNA 21: 840–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Singh ND, Li L, Zhang X & Daniell H (2015) Engineered chloroplast dsRNA silences cytochrome p450 monooxygenase, V‐ATPase and chitin synthase genes in the insect gut and disrupts Helicoverpa armigera larval development and pupation. Plant Biotechnology Journal 13: 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose AM & Hunter CP (2007) Transport of sequence‐specific RNA interference information between cells. Annual Review of Genetics 41: 305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose AM, Smith JJ & Hunter CP (2009) Export of RNA silencing from C. elegans tissues does not require the RNA channel SID‐1. Proceedings of the National Academy of Sciences of the USA 106: 2283–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose AM, Kim YA, Leal‐Ekman S & Hunter CP (2012) Conserved tyrosine kinase promotes the import of silencing RNA into Caenorhabditis elegans cells. Proceedings of the National Academy of Sciences of the USA 109: 14520–14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph TT & Osman R (2012) Convergent transmission of RNAi guide‐target mismatch information across Argonaute internal allosteric network. PLoS Computational Biology 8: 1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez‐Campos M, Zipperlen P, Fraser AG & Ahringer J (2001) Effectiveness of specific RNA‐mediated interference through ingested double‐stranded RNA in Caenorhabditis elegans . Genome Biology 2: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S, Troiano E & Mather TN (2010) Functional genomics tool: gene silencing in Ixodes scapularis eggs and nymphs by electroporated dsRNA. BMC Biotechnology 10: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajuria C, Vélez AM, Rangasamy M, Wang H, Fishilevich E et al. (2015) Parental RNA interference of genes involved in embryonic development of the western corn rootworm, Diabrotica virgifera virgifera LeConte. Insect Biochemistry and Molecular Biology 63: 54–62. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A & Jayasena SD (2003) Functional siRNAs and rniRNAs exhibit strand bias. Cell 115: 209–216. [DOI] [PubMed] [Google Scholar]
- Killiny N, Hajeri S, Tiwari S, Gowda S & Stelinski LL (2014) Double‐stranded RNA uptake through topical application, mediates silencing of five CYP4 genes and suppresses insecticide resistance in Diaphorina citri . PLoS ONE 9: e110536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J & Siomi MC (2009) Biogenesis of small RNAs in animals. Nature Reviews Molecular Cell Biology 10: 126–139. [DOI] [PubMed] [Google Scholar]
- Kim YH, Issa MS, Cooper AMW & Zhu KY (2015) RNA interference: applications and advances in insect toxicology and insect pest management. Pesticide Biochemistry and Physiology 120: 109–117. [DOI] [PubMed] [Google Scholar]
- Knipling EF (1955) Possibilities of insect control or eradication through the use of sexually sterile males. Journal of Economic Entomology 48: 459–462. [Google Scholar]
- Knipling EF (1998) Role of parasitoid augmentation and sterile insect techniques for area‐wide management of agricultural insect pests. Journal of Agricultural Entomology 15: 273–301. [Google Scholar]
- Krafsur ES (1998) Sterile insect technique for suppressing and eradicating insect populations: 55 years and counting. Journal of Agricultural Entomology 15: 303–317. [Google Scholar]
- Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P et al. (2006) Evidence of off‐target effects associated with long dsRNAs in Drosophila melanogaster cell‐based assays. Nature Methods 3: 833–838. [DOI] [PubMed] [Google Scholar]
- Kumar P, Pandit SS & Baldwin IT (2012) Tobacco rattle virus vector: a rapid and transient means of silencing Manduca sexta genes by plant mediated RNA interference. PLoS ONE 7: e31347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi‐Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y et al. (2001) Two mouse piwi‐related genes: miwi and mili. Mechanisms of Development 108: 121–133. [DOI] [PubMed] [Google Scholar]
- Lan H, Chen H, Liu Y, Jiang C, Mao Q et al. (2016a) Small interfering RNA pathway modulates initial viral infection in midgut epithelium of insect after ingestion of virus. Journal of Virology 90: 917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan H, Wang H, Chen Q, Chen H, Jia D et al. (2016b) Small interfering RNA pathway modulates persistent infection of a plant virus in its insect vector. Scientific Reports 6: 20699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P‐W, Potter CS, Carragher B & MacRae IJ (2009) Structure of the human Dicer‐TRBP complex by electron microscopy. Structure 17: 1326–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z et al. (2004) Distinct roles for Drosophila Dicer‐1 and Dicer‐2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81. [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR & Kim VN (2006) The role of PACT in the RNA silencing pathway. EMBO Journal 25: 522–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HY, Zhou K, Smith AM, Noland CL & Doudna JA (2013) Differential roles of human Dicer‐binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Research 41: 6568–6576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner PJF, Ameres SL, Kueng S & Martinez J (2006) Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Reports 7: 314–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang M & Zhang H (2011) RNA interference of four genes in adult Bactrocera dorsalis by feeding their dsRNAs. PLoS ONE 6: e0017788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li X, Bai R, Shi Y, Tang Q et al. (2015a) RNA interference of the P450 CYP6CM1 gene has different efficacy in B and Q biotypes of Bemisia tabaci . Pest Management Science 71: 1175–1181. [DOI] [PubMed] [Google Scholar]
- Li X, Dong X, Zou C & Zhang H (2015b) Endocytic pathway mediates refractoriness of insect Bactrocera dorsalis to RNA interference. Scientific Reports 5: 08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dou K, Gao S, Sun J, Wang M et al. (2015c) Impacts on silkworm larvae midgut proteomics by transgenic Trichoderma strain and analysis of glutathione S‐transferase sigma 2 gene essential for anti‐stress response of silkworm larvae. Journal of Proteomics 126: 218–227. [DOI] [PubMed] [Google Scholar]
- Li Z, Zeng B, Ling L, Xu J, You L et al. (2015d) Enhancement of larval RNAi efficiency by over‐expressing Argonaute2 in Bombyx mori . International Journal of Biological Sciences 11: 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Wang Y, Murota Y, Liu X, Smith D et al. (2015) TAF11 assembles the RISC loading complex to enhance RNAi efficiency. Molecular Cell 59: 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Li W, Zhang T, Kirberger M, Liu J et al. (2016) Powering up the molecular therapy of RNA interference by novel nanoparticles. Biomaterials Science 4: 1051–1061. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM et al. (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441. [DOI] [PubMed] [Google Scholar]
- Liu X, Jiang F, Kalidas S, Smith D & Liu Q (2006) Dicer‐2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA 12: 1514–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ye X, Jiang F, Liang C, Chen D et al. (2009) C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science 325: 750–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Swevers L, Iatrou K, Huvenne H & Smagghe G (2012) Bombyx mori DNA/RNA non‐specific nuclease: expression of isoforms in insect culture cells, subcellular localization and functional assays. Journal of Insect Physiology 58: 1166–1176. [DOI] [PubMed] [Google Scholar]
- Liu G, Wu Q, Li J, Zhang G & Wan F (2015) RNAi‐mediated knock‐down of transformer and transformer 2 to generate male‐only progeny in the oriental fruit fly, Bactrocera dorsalis (Hendel). PLoS ONE 10: e0128892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Zhou Y, Yu Y, Cao J, Zhang H et al. (2015) RNA interference and the vaccine effect of a subolesin homolog from the tick Rhipicephalus haemaphysaloides . Experimental and Applied Acarology 68: 113–126. [DOI] [PubMed] [Google Scholar]
- Luo KQ & Chang DC (2004) The gene‐silencing efficiency of siRNA is strongly dependent on the local structure of mRNA at the targeted region. Biochemical and Biophysical Research Communications 318: 303–310. [DOI] [PubMed] [Google Scholar]
- Ma Y, Creanga A, Lum L & Beachy PA (2006) Prevalence of off‐target effects in Drosophila RNA interference screens. Nature 443: 359–363. [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Ma E, Zhou M, Robinson CV & Doudna JA (2008) In vitro reconstitution of the human RISC‐loading complex. Proceedings of the National Academy of Sciences of the USA 105: 512–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HJ, Raza A, Amin I, Scheffler JA, Scheffler BE et al. (2016) RNAi‐mediated mortality of the whitefly through transgenic expression of double‐stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Scientific Reports 6: 38469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR et al. (2009) Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamta B, Reddy KRK & Rajam MV (2015) Targeting chitinase gene of Helicoverpa armigera by host‐induced RNA interference confers insect resistance in tobacco and tomato. Plant Molecular Biology 90: 281–292. [DOI] [PubMed] [Google Scholar]
- Maniataki E & Mourelatos Z (2005) A human, ATP‐independent, RISC assembly machine fueled by pre‐miRNA. Genes and Development 19: 2979–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J & Zeng F (2012) Feeding‐based RNA intereference of a gap gene is lethal to the pea aphid, Acyrthosiphon pisum . PLoS ONE 7: e48718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP & Zamore PD (2005) Passenger‐strand cleavage facilitates assembly of siRNA into Ago2‐containing RNAi enzyme complexes. Cell 123: 607–620. [DOI] [PubMed] [Google Scholar]
- Mayer DG, Atzeni MG, Stuart MA, Anaman KA & Butler DG (1998) Mating competitiveness of irradiated flies for screwworm fly eradication campaigns. Preventive Veterinary Medicine 36: 1–9. [DOI] [PubMed] [Google Scholar]
- McEwan D, Weisman A & Hunter CP (2012) Uptake of extracellular double‐stranded RNA by SID‐2. Molecular Cell 47: 746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SC, Miyata K, Brown SJ & Tomoyasu Y (2012) Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: parameters affecting the efficiency of RNAi. PLoS ONE 7: e47431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata K, Ramaseshadri P, Zhang Y, Segers G, Bolognesi R & Tomoyasu Y (2014) Establishing an in vivo assay system to identify components involved in environmental RNA interference in the western corn rootworm. PLoS ONE 9: e101661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN & Myles KM (2012) Production of virus‐derived ping‐pong‐dependent piRNA‐like small RNAs in the mosquito soma. PLoS Pathogens 8: e1002470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Béclin C, Boutet S et al. (2002) Fertile hypomorphic Argonaute (ago1) mutants impaired in post‐transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S & Hannon GJ (2005) Characterization of Dicer‐deficient murine embryonic stem cells. Proceedings of the National Academy of Sciences of the USA 102: 12135–12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore K, Sun L, Tomchaney M, Sullivan G, Adams H et al. (2015) siRNA‐mediated silencing of doublesex during female development of the dengue vector mosquito Aedes aegypti . PLoS Neglected Tropical Diseases 9: e4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Yamada T, Matsumiya T, Ui‐Tei K, Saigo K & Morishita S (2005) dsCheck: highly sensitive off‐target search software for double‐stranded RNA‐mediated RNA interference. Nucleic Acids Research 33: W589–W591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C, Lemieux C & Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co‐suppression of homologous genes in trans. Plant Cell 2: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland CL & Doudna JA (2013) Multiple sensors ensure guide strand selection in human RNAi pathways. RNA 19: 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland CL, Ma E & Doudna JA (2011) siRNA repositioning for guide strand selection by human Dicer complexes. Molecular Cell 43: 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP & Lai EC (2008) The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453: 803–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban TI & Izaurralde E (2005) Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 11: 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paim RMM, Araujo RN, Lehane MJ, Gontijo NF & Pereira MH (2013) Long‐term effects and parental RNAi in the blood feeder Rhodnius prolixus (Hemiptera; Reduviidae). Insect Biochemistry and Molecular Biology 43: 1015–1020. [DOI] [PubMed] [Google Scholar]
- Pak J & Fire A (2007) Distinct populations of primary and secondary effectors during RNAi in C. elegans . Science 315: 241–244. [DOI] [PubMed] [Google Scholar]
- Pan ZH, Gao K, Hou CX, Wu P, Qin GX et al. (2016) Corrigendum to ‘dsRNA interference on expression of a RNA‐dependent RNA polymerase gene of Bombyx mori cytoplasmic polyhedrosis virus’ [Gene (2015) 565: 56–61]. Gene 586: 282. [DOI] [PubMed] [Google Scholar]
- Pane A, Salvemini M, Bovi PD, Polito C & Saccone G (2002) The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 129: 3715–3725. [DOI] [PubMed] [Google Scholar]
- Peng W, Zheng W, Handler AM & Zhang H (2015) The role of the transformer gene in sex determination and reproduction in the tephritid fruit fly, Bactrocera dorsalis (Hendel). Genetica 143: 717–727. [DOI] [PubMed] [Google Scholar]
- Pridgeon JW, Zhao L, Becnel JJ, Strickman DA, Clark GG & Linthicum KJ (2008) Topically applied AaeIAP1 double‐stranded RNA kills female adults of Aedes aegypti . Journal of Medical Entomology 45: 414–420. [DOI] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du FH & Wang XD (2005) Argonaute2 cleaves the anti‐guide strand of siRNA during RISC activation. Cell 123: 621–629. [DOI] [PubMed] [Google Scholar]
- Ratzka C, Gross R & Feldhaar H (2013) Systemic gene knockdown in Camponotus floridanus workers by feeding of dsRNA. Insectes Sociaux 60: 475–484. [Google Scholar]
- Rebijith KB, Asokan R, Hande HR, Kumar NKK, Krishna V et al. (2016) RNA interference of odorant‐binding protein 2 (OBP2) of the cotton aphid, Aphis gossypii (Glover), resulted in altered electrophysiological responses. Applied Biochemistry and Biotechnology 178: 251–266. [DOI] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS & Khvorova A (2004) Rational siRNA design for RNA interference. Nature Biotechnology 22: 326–330. [DOI] [PubMed] [Google Scholar]
- Ruiz N, De Abreu LA, Parizi LF, Kim TK, Mulenga A et al. (2015) Non‐invasive delivery of dsRNA into de‐waxed tick eggs by electroporation. PLoS ONE 10: e130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V et al. (2009) Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature 458: 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvemini M, Robertson M, Aronson B, Atkinson P, Polito LC & Saccone G (2009) Ceratitis capitata transformer‐2 gene is required to establish and maintain the autoregulation of Cctra, the master gene for female sex determination. International Journal of Developmental Biology 53: 109–120. [DOI] [PubMed] [Google Scholar]
- Sandoval‐Mojica AF & Scharf ME (2016) Silencing gut genes associated with the peritrophic matrix of Reticulitermes flavipes (Blattodea: Rhinotermitidae) increases susceptibility to termiticides. Insect Molecular Biology 25: 734–744. [DOI] [PubMed] [Google Scholar]
- Schnettler E, Donald CL, Human S, Watson M, Siu RW et al. (2013) Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. Journal of General Virology 94: 1680–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N & Zamore PD (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208. [DOI] [PubMed] [Google Scholar]
- Scott JG, Michel K, Bartholomay LC, Siegfried BD, Hunter WB et al. (2013) Towards the elements of successful insect RNAi. Journal of Insect Physiology 59: 1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JK, Lee GS, Lee S & Lee KY (2015) Oral ingestion of heat shock protein 70 dsRNA is lethal under normal and thermal stress conditions in the sweetpotato whitefly, Bemisia tabaci . Journal of Asia‐Pacific Entomology 18: 797–800. [Google Scholar]
- Shukla JN & Palli SR (2012) Sex determination in beetles: production of all male progeny by Parental RNAi knockdown of transformer. Scientific Reports 2: 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla JN, Kalsi M, Sethi A, Narva KE, Fishilevich E et al. (2016) Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biology 13: 656–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S et al. (2001) On the role of RNA amplification in dsRNA‐triggered gene silencing. Cell 107: 465–476. [DOI] [PubMed] [Google Scholar]
- Singh AD, Wong S, Ryan CP & Whyard S (2013) Oral delivery of double‐stranded RNA in larvae of the yellow fever mosquito, Aedes aegypti: implications for pest mosquito control. Journal of Insect Science 13: 6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ & Joshua‐Tor L (2004) Crystal structure of argonaute and its implications for RISC slicer activity. Science 305: 1434–1437. [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ (2005) Assembly and function of RNA silencing complexes. Nature Reviews Molecular Cell Biology 6: 127–138. [DOI] [PubMed] [Google Scholar]
- Sørensen JG, Addison MF & Terblanche JS (2012) Mass‐rearing of insects for pest management: challenges, synergies and advances from evolutionary physiology. Crop Protection 38: 87–94. [Google Scholar]
- Sugahara R, Tanaka S, Jouraku A & Shiotsuki T (2017) Geographic variation in RNAi sensitivity in the migratory locust. Gene 605: 5–11. [DOI] [PubMed] [Google Scholar]
- Swevers L, Huvenne H, Menschaert G, Kontogiannatos D, Kourti A et al. (2013) Colorado potato beetle (Coleoptera) gut transcriptome analysis: expression of RNA interference‐related genes. Insect Molecular Biology 22: 668–684. [DOI] [PubMed] [Google Scholar]
- Tabashnik BE, Mota‐Sanchez D, Whalon ME, Hollingworth RM & Carriere Y (2014) Defining terms for proactive management of resistance to Bt crops and pesticides. Journal of Economic Entomology 107: 496–507. [DOI] [PubMed] [Google Scholar]
- Tabor S (2001) Expression using the T7 RNA polymerase/promoter system. Current Protocols in Molecular Biology Unit16.2; https://doi.org/10.1002/0471142727.mb1602s11 [DOI] [PubMed] [Google Scholar]
- Taning CNT, Christiaens O, Berkvens N, Casteels H, Maes M & Smagghe G (2016) Oral RNAi to control Drosophila suzukii: laboratory testing against larval and adult stages. Journal of Pest Science 89: 803–814. [Google Scholar]
- Taracena ML, Oliveira PL, Almendares O, Umaña C, Lowenberger C et al. (2015) Genetically modifying the insect gut microbiota to control Chagas disease vectors through systemic RNAi. PLoS Neglected Tropical Diseases 9: e3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DD, Donnelly CA, Wood RJ & Alphey LS (2000) Insect population control using a dominant, repressible, lethal genetic system. Science 287: 2474. [DOI] [PubMed] [Google Scholar]
- Tian H, Peng H, Yao Q, Chen H, Xie Q et al. (2009) Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non‐midgut gene. PLoS ONE 4: e6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Cheng L, Qi X, Ge Z, Niu C et al. (2015) Transgenic cotton plants expressing double‐stranded RNAs target HMG‐CoA reductase (HMGR) gene inhibits the growth, development and survival of cotton bollworms. International Journal of Biological Sciences 11: 1296–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo J, Rull J, Oropeza A, Hernandez E & Liedo P (2004) Irradiation of Anastrepha obliqua (Diptera: Tephritidae) revisited: optimizing sterility induction. Journal of Economic Entomology 97: 383–389. [DOI] [PubMed] [Google Scholar]
- Tomari Y & Zamore PD (2005) Machines for RNAi. Genes and Development 19: 517–529. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N & Zamore PD (2004) A protein sensor for siRNA asymmetry. Science 306: 1377–1380. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T & Zamore PD (2007) Sorting of Drosophila small silencing RNAs. Cell 130: 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu Y, Miller SC, Tomita S, Schoppmeier M, Grossmann D & Bucher G (2008) Exploring systemic RNA interference in insects: a genome‐wide survey for RNAi genes in Tribolium . Genome Biology 9: R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toprak U, Baldwin D, Erlandson M, Gillott C, Harris S & Hegedus DD (2013) In vitro and in vivo application of RNA interference for targeting genes involved in peritrophic matrix synthesis in a lepidopteran system. Insect Science 20: 92–100. [DOI] [PubMed] [Google Scholar]
- Tzin V, Yang X, Jing X, Zhang K, Jander G & Douglas AE (2015) RNA interference against gut osmoregulatory genes in phloem‐feeding insects. Journal of Insect Physiology 79: 105–112. [DOI] [PubMed] [Google Scholar]
- Ulrich J, Dao VA, Majumdar U, Schmitt‐Engel C, Schwirz J et al. (2015) Large scale RNAi screen in Tribolium reveals novel target genes for pest control and the proteasome as prime target. BMC Genomics 16: 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V & Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324. [DOI] [PubMed] [Google Scholar]
- Vaucheret H (2006) Post‐transcriptional small RNA pathways in plants: mechanisms and regulations. Genes and Development 20: 759–771. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S, Tuschl T & Patel DJ (2008) Structure of the guide‐strand‐containing argonaute silencing complex. Nature 456: 209–U234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Li S, Qi HH, Chowdhury D, Shi Y & Novina CD (2009) Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nature Structural and Molecular Biology 16: 1259–U1276. [DOI] [PubMed] [Google Scholar]
- Wei Q, Wu SF, Niu CD, Yu HY, Dong YX & Gao CF (2015) Knockdown of the ionotropic γ‐aminobutyric acid receptor (gabar) rdl gene decreases fipronil susceptibility of the small brown planthopper, Laodelphax striatellus (Hemiptera: Delphacidae). Archives of Insect Biochemistry and Physiology 88: 249–261. [DOI] [PubMed] [Google Scholar]
- Whangbo JS & Hunter CP (2008) Environmental RNA interference. Trends in Genetics 24: 297–305. [DOI] [PubMed] [Google Scholar]
- Whitten MMA, Facey PD, del Sol R, Fernández‐Martínez LT, Evans MC et al. (2015) Symbiont‐mediated RNA interference in insects. Proceedings of the Royal Society B 283: 20160042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyard S, Singh AD & Wong S (2009) Ingested double‐stranded RNAs can act as species‐specific insecticides. Insect Biochemistry and Molecular Biology 39: 824–832. [DOI] [PubMed] [Google Scholar]
- Whyard S, Erdelyan CN, Partridge AL, Singh AD, Beebe NW & Capina R (2015) Silencing the buzz: a new approach to population suppression of mosquitoes by feeding larvae double‐stranded RNAs. Parasites and Vectors 8: 716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C & Hunter CP (2002) Systemic RNAi in C. elegans requires the putative transmembrane protein SID‐1. Science 295: 2456–2459. [DOI] [PubMed] [Google Scholar]
- Winston WM, Sutherlin M, Wright AJ, Feinberg EH & Hunter CP (2007) Caenorhabditis elegans SID‐2 is required for environmental RNA interference. Proceedings of the National Academy of Sciences of the USA 104: 10565–10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynant N, Santos D, Van Wielendaele P & Vanden Broeck J (2014) Scavenger receptor‐mediated endocytosis facilitates RNA interference in the desert locust, Schistocerca gregaria . Insect Molecular Biology 23: 320–329. [DOI] [PubMed] [Google Scholar]
- Xiao D, Gao X, Xu J, Liang X, Li Q et al. (2015) Clathrin‐dependent endocytosis plays a predominant role in cellular uptake of double‐stranded RNA in the red flour beetle. Insect Biochemistry and Molecular Biology 60: 68–77. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Zeng H, Zhang Y, Xu D & Qiu D (2013) Silencing the HaHR3 gene by transgenic plant‐mediated RNAi to disrupt Helicoverpa armigera development. International Journal of Biological Sciences 9: 370–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Feng HT, Wang C, Yip KH, Pavlos N et al. (2003) Effects of Bafilomycin A1: an inhibitor of vacuolar H (+)‐ATPases on endocytosis and apoptosis in RAW cells and RAW cell‐derived osteoclasts. Journal of Cellular Biochemistry 88: 1256–1264. [DOI] [PubMed] [Google Scholar]
- Xue J, Ye Y‐X, Jiang Y‐Q, Zhuo JIC, Huang H‐J et al. (2015) Efficient RNAi of rice planthoppers using microinjection. Protocol Exchange. https://doi.org/10.1038/protex.2015.005. [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L & Zhou MM (2003) Structure and conserved RNA binding of the PAZ domain. Nature 426: 469–474. [DOI] [PubMed] [Google Scholar]
- Yang J & Han Z‐J (2014) Efficiency of different methods for dsRNA delivery in cotton bollworm (Helicoverpa armigera). Journal of Integrative Agriculture 13: 115–123. [Google Scholar]
- Ye X, Huang N, Liu Y, Paroo Z, Huerta C et al. (2011) Structure of C3PO and mechanism of human RISC activation. Nature Structural & Molecular Biology 18: 650–U643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Zhao H, Wang P, Hu M & Zhong G (2014) Bdor\Orco is important for oviposition‐deterring behavior induced by both the volatile and non‐volatile repellents in Bactrocera dorsalis (Diptera: Tephritidae). Journal of Insect Physiology 65: 51–56. [DOI] [PubMed] [Google Scholar]
- Yoon JS, Shukla JN, Gong ZJ, Mogilicherla K & Palli SR (2016) RNA interference in the Colorado potato beetle, Leptinotarsa decemlineata: identification of key contributors. Insect Biochemistry and Molecular Biology 78: 78–88. [DOI] [PubMed] [Google Scholar]
- Yu R, Xu X, Liang Y, Tian H, Pan Z et al. (2014) The insect ecdysone receptor is a good potential target for RNAi‐based pest control. International Journal of Biological Sciences 10: 1171–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Li DT, Wang SL, Xu HJ, Bao YY & Zhang CX (2016) Ion transport peptide (ITP) regulates wing expansion and cuticle melanism in the brown planthopper, Nilaparvata lugens . Insect Molecular Biology 25: 778–787. [DOI] [PubMed] [Google Scholar]
- Zhang HD, Kolb FA, Brondani V, Billy E & Filipowicz W (2002) Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO Journal 21: 5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang J & Zhu KY (2010) Chitosan/double‐stranded RNA nanoparticle‐mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae). Insect Molecular Biology 19: 683–693. [DOI] [PubMed] [Google Scholar]
- Zhang J, Khan SA, Hasse C, Ruf S, Heckel DG & Bock R (2015a) Full crop protection from an insect pest by expression of long double‐stranded RNAs in plastids. Science 347: 991–994. [DOI] [PubMed] [Google Scholar]
- Zhang X, Mysore K, Flannery E, Michel K, Severson DW et al. (2015b) Chitosan/interfering RNA nanoparticle mediated gene silencing in disease vector mosquito larvae. Journal of Visualized Experiments 97: e52523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Liu Y, Zheng W, Xiao Y & Zhang H (2015) Influence of the silencing sex‐peptide receptor on Bactrocera dorsalis adults and offspring by feeding with ds‐spr. Journal of Asia‐Pacific Entomology 18: 477–481. [Google Scholar]
- Zhou X, Wheeler MM, Oi FM & Scharf ME (2008) RNA interference in the termite Reticulitermes flavipes through ingestion of double‐stranded RNA. Insect Biochemistry and Molecular Biology 38: 805–815. [DOI] [PubMed] [Google Scholar]
- Zhu F, Xu J, Palli R, Ferguson J & Palli SR (2011) Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata . Pest Management Science 67: 175–182. [DOI] [PubMed] [Google Scholar]
- Zhu JQ, Liu S, Ma Y, Zhang JQ, Qi HS et al. (2012) Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect‐associated gene EcR. PLoS ONE 7: e38572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwier MV, Verhulst EC, Zwahlen RD, Beukeboom LW & van de Zande L (2012) DNA methylation plays a crucial role during early Nasonia development. Insect Molecular Biology 21: 129–138. [DOI] [PubMed] [Google Scholar]


