Figure 3.
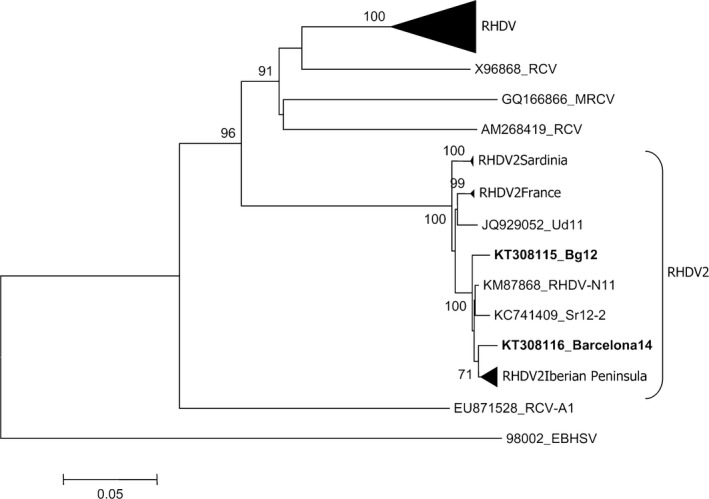
Phylogenetic analysis of VP60 sequences of RHDV2 strains identified in hares in Italy and Spain. For the phylogenetic analysis of the VP60 sequences, the neighbour‐joining method was applied based on the Kimura 2 parameters using the software package MEGA6 (Tamura et al., 2013). The European brown hare syndrome virus (EBHSV) strain BS89 (GenBank Accession No.: X98002) was used as an out‐group to root the tree. Bootstrap probability values above 70% for 1000 replicate runs are indicated at the nodes. Twenty‐one selected sequences representing previously described RHDV genogroups (G1–G6; GenBank a.n.: EF558575, Y15441, X87607, Y15426, AJ535094, AJ535092, Y15427, AJ495856, KC595270, KC345614, Z49271, Z29514, Y15440, L48547, AF402614, U49726, M67473, U54983, Y15425, EU250330, AJ302016), the sequences of the RHDV2 isolates identified in France (GenBank a.n.: HE819400, HE800530, HE819400), in Sardinia/Italy (GenBank Accession No.: KC345611, KC345613, JX106023, KC345612) (2010–2012) and in the Iberian Peninsula (GenBank a.n.: KM115712, KM115713, KM115714, KM115715, KM115716, KP129397, KM115697, KM115689) (2012–2014) have been collapsed. The accession numbers are indicated for the remaining RHDV2 sequences, including RHDV‐N11 (GenBank a.n.: KM87868), the first Spanish RHDV2 isolate; RHDV2_Sr12‐2 strain (GenBank a.n.: KC741409), the isolate from Italian hares in Sicily; and Ud11 (GenBank a.n.: JQ929052), the first RHDV2 isolated in Italy. The sequences of the Spanish (Barcelona14, GenBank a.n.: KT308116) and Italian (Bg12, GenBank a.n.: KT308115) RHDV2 hare strains are in bold. Four sequences of European (X96868_RCV, Italy 1995; GQ166866_MRCV, USA 2001; AM268419_RCV, France 2006) and Australian (EU871528_RCVA1) non‐pathogenic RCVs were also included.
