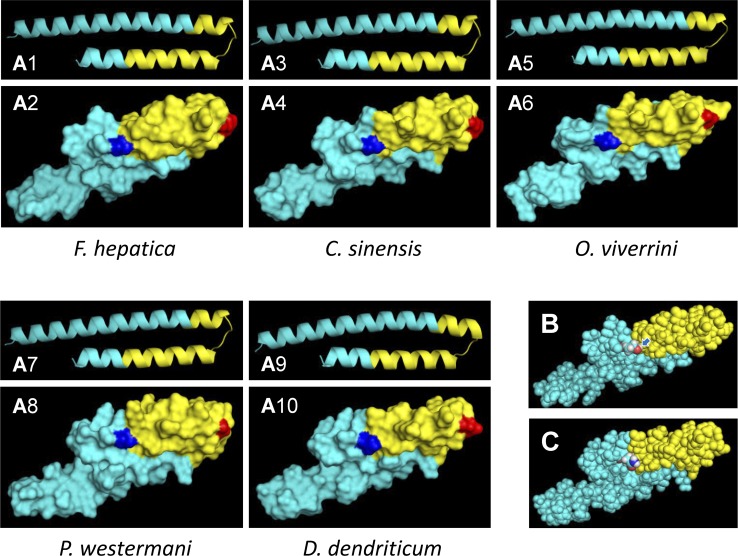
Fig 5. Three-dimensional (3D) structure predictions of MF6p/FhHDM-1 orthologs from different trematodes.
A) Predicted PDB structures for MF6p/FhHDM-1 orthologs from F. hepatica, C. sinensis, O. viverrini, P. westermani and D. dendriticum using the Quark Online ab initio protein folding and protein structure prediction bioinformatics tool. For better visualization, each 3D structure was represented as a cartoon (odd numbers) or as a surface (even numbers). The overall surface structures are shown in cyan, the area corresponding to the MF6-recognized epitope is represented in yellow and the positions corresponding to 64N and 81T in the sequence of F. hepatica, assumed to be highly relevant for MF6 epitope conformation, are shown in red and blue, respectively. B) Theoretical 3D structure of the MF6p/FhHDM-1 mature protein (represented as balls) showing the overall structure of the MF6-recognized epitope (shaded in yellow) and the position of the N-terminal 81T residue (colored atoms; red = oxygen; white = hydrogen; cyan = carbon). The arrow shows the position of the hydroxyl group of the 81T residue. C) Theoretical 3D structure of the MF6p/FhHDM-1 mature protein ortholog from D. dendriticum (represented as balls) highlighting the equivalent region to the MF6-recognized epitope (shaded in yellow) and the position of the N-terminal 81N residue in substitution of 81T in F. hepatica (colored atoms; red = oxygen, see arrow; white = hydrogen; cyan = carbon). All 3D representations of the PDB models obtained with the Quark Online bioinformatics tool were elaborated using the PyMOL Molecular Graphics System.