Summary
Pectate lyase genes have been documented as excellent candidates for improvement of fruit firmness. However, implementation of pectate lyase in regulating fruit postharvest deterioration has not been fully explored. In this report, 22 individual pectate lyase genes in tomato were identified, and one pectate lyase gene SlPL (Solyc03g111690) showed dominant expression during fruit maturation. RNA interference of SlPL resulted in enhanced fruit firmness and changes in pericarp cells. More importantly, the SlPL‐RNAi fruit demonstrated greater antirotting and pathogen‐resisting ability. Compared to wild‐type, SlPL‐RNAi fruit had higher levels of cellulose and hemicellulose, whereas the level of water‐soluble pectin was lower. Consistent with this, the activities of peroxidase, superoxide dismutase and catalase were higher in SlPL‐RNAi fruit, and the malondialdehyde concentration was lower. RNA‐Seq results showed large amounts of differentially expressed genes involved in hormone signalling, cell wall modification, oxidative stress and pathogen resistance. Collectively, these data demonstrate that pectate lyase plays an important role in both fruit softening and pathogen resistance. This may advance knowledge of postharvest fruit preservation in tomato and other fleshy fruit.
Keywords: pectate lyase, SlPL, RNA interference, fruit shelf‐life, pathogen resistance, tomato
Introduction
Postharvest deterioration is one of the biggest challenges for vegetable and fruit industries (Meli et al., 2010). The main determinant for the postharvest deterioration of fruit is the rate of softening, which limits fruit shelf‐life and affects wastage, postharvest pathogen‐induced infection and frequency of harvest (Brummell and Harpster, 2001). Hence, engineering fruit goods with better resistance to postharvest losses and diseases possesses potential value. Fruit softening is a complicated process regulated by a multitude of factors, among which cell wall modifying enzymes are considered to be the main contributors (Lunn et al., 2013). The plant cell wall is a highly complex matrix containing hemicellulose, cellulose, pectins and other components (Carpita and Gibeaut, 1993; Cassab and Varner, 1988; Tieman and Handa, 1994). As the ripening process begins, a series of biochemical changes occur at the cell wall resulting in the breakdown of cell wall polymers (Rose et al., 1997). The disintegration of cell walls can then lead to a reduction in intercellular adhesion, solubilization of pectins, depolymerization of hemicelluloses and loss of pectic galactose side chains.
Pectin is a main component of the primary cell wall, which plays a key role in maintaining the physical structure stability and mechanical strength of the cell wall. Galacturonic acid (GalA) is the major constituent of pectin, with a weight ratio of 70%. Pectin polysaccharides primarily comprise homogalacturonan (HG), xylogalacturonan (XGA), rhamnogalacturonan II (RG‐II) and rhamnogalacturonan I (RG‐I). HG is a linear polymer of the α‐1,4‐linked GalA, and XGA is a modified HG. In RG‐II, the HG backbone is complemented by lateral chains composed of 12 different sugars attached by 22 distinct linkages. The backbone of RG‐I is essentially a disaccharide repeating unit (α‐1,4‐D‐GalA‐α‐1,2‐L‐Rha) where the L‐rhamnose (Rha) can be branched with arabinan, galactan or arabinogalactan (Harholt et al., 2010). Because of the complex composition and spatial structure of pectic substances, many enzymes are required to catalyse the fruit softening process (Huber, 1984; Knee et al., 1977; Redgwell et al., 1997). Polygalacturonase (PG) and pectin methylesterase (PME) are the two most thoroughly characterized pectin‐modifying enzymes, but they are incapable of significantly affecting fruit texture (Grierson and Schuch, 1993; Hall et al., 1993). In tomato (Solanum lycopersicum), inhibition of PG activity to 1% level did not prevent pectin solubilization (Langley et al., 1994; Sheehy et al., 1988; Smith et al., 1990), and antisense inhibition of PME genes had relatively little effect on fruit softening (Tieman et al., 1992; Wen et al., 2013). Similar results were obtained in studies associated with endo‐glucanase (Brummell et al., 1999) and xyloglucan endotransglycosylase (De Silva et al., 1994). Additionally, suppressing other genes such as expansin and β‐galactosidase 4 had a moderate effect on regulating the softening process, but the changes were modest at most (Brummell et al., 1999, 2002; Smith et al., 2002).
Pectate lyases (PLs) (EC 4.2.2.2) are one kind of pectin‐modifying enzymes that are capable of cleaving glycosidic bonds through a β‐elimination mechanism between galacturonosyl residues (Marín‐Rodríguez et al., 2002; Willats et al., 2001). PL‐like sequences from higher plants were first reported in pollen (Wing et al., 1989). It was suggested that PL genes were required for the initial loosening of pollen cell walls to enable pollen tube growth and facilitate penetration of pollen (Taniguchi et al., 1995; Wu et al., 1996). PL sequences have been reported in climacteric fruit including banana (Domínguez ‐Puigjaner et al., 1997; Marín‐Rodríguez, 2001; Medina‐Suárez et al., 1997; Pilatzke‐Wunderlich and Nessler, 2001) and mango (Chourasia et al., 2006), as well as in nonclimacteric fruit including strawberry (Medina‐Escobar et al., 1997) and grape berries (Nunan et al., 2001). PL gene expression was first manipulated with transgenic approaches in strawberry, where the suppression of PL mRNA during ripening resulted in significantly firmer fruit (Jiménez‐Bermúdez et al., 2002; Santiago‐Doménech et al., 2008). Very recently, Uluisik et al. (2016) showed that silencing a PL gene in tomato led to targeted control of tomato softening, without affecting other aspects of ripening.
Tomato is one of the most important agricultural products in the world, as well as an important model for fleshy fruit ripening (Giovannoni, 2001). To further analyse the roles of pectate lyase in tomato, we identified 22 gene family members via in silico mining of the tomato genome (http://solgenomics.net/). One fruit softening‐specific gene Solyc03g111690 named SlPL in accordance with the recent study was cloned, and its functions were characterized through RNA interference (RNAi)‐mediated gene silencing. Silencing SlPL in tomato led to enhanced fruit firmness, extended fruit shelf‐life and reduced susceptibility to Botrytis cinerea. The findings indicated that SlPL could be a useful biotechnological tool in genetic engineering of fleshy fruit.
Results
Identification, sequence analysis and expression profiles of tomato PL genes
By mining the annotated tomato genome database (SOL Genomics Network, http://solgenomics.net/), we identified 22 tomato PL genes. A phylogenetic analysis of tomato PL sequences, together with all 26 Arabidopsis thaliana PLs and two softening‐related PLs from strawberry and banana (Data S1), was carried out using the neighbour‐joining method on MEGA6 to investigate the relationship between SlPLs and other PLs. The analysis revealed that the tomato genome carried all genes orthologous to those described previously in Arabidopsis and the tomato PL proteins could be divided into five subfamilies (Figure 1a), consistent with a previous investigation by Singh et al. (2011). Alignment of predicted PL domain sequences showed that three typical motifs, motif 1 (WIDH), motif 2 (DGLIDAIMASTAITISNNYF) and motif 3 (LIQRMPRCRHGYFHVVNNDY) could be recognized in the sequence (Figure 1b), thus further confirming the identity of the predicted PLs in tomato. Next, comprehensive transcriptomic profiling of 22 tomato PLs in vegetative and reproductive tissues was carried out using the online TomExpress platform and associated data‐mining tools (http://gbf.toulouse.inra.fr/tomexpress) (Figure 1c). Notably, we found that Solyc03 g111690 (hereafter designated as SlPL) had higher expression during fruit ripening and softening than other gene family members, suggesting that this gene might play important roles in regulating the fruit ripening/softening process.
Figure 1.
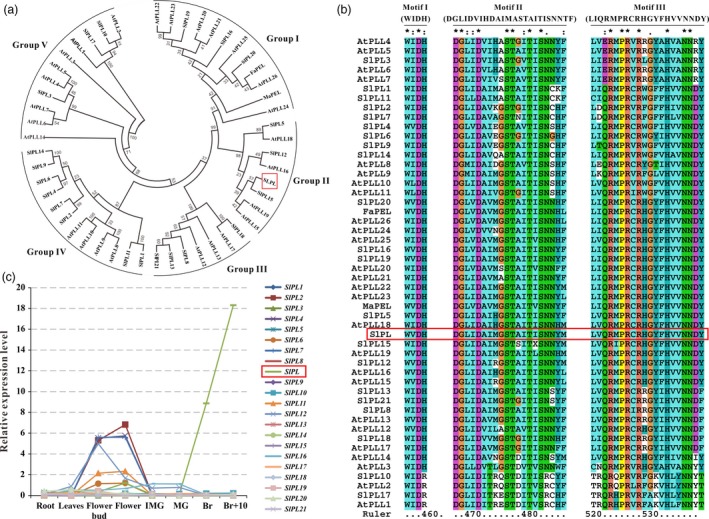
Phylogenetic analysis, multiple sequence alignment and expression patterns of pectate lyases (PLs). (a) Phylogenetic analysis of tomato PL gene family members, together with all 26 Arabidopsis PLs and two softening‐related PLs from strawberry (FaPEL) and banana (MaPEL), all of which were carried out by the neighbour‐joining method on MEGA6. (b) Three typical motifs (motif 1 WIDH, motif 2 DGLIDAIMASTAITISNNYF and motif 3 LIQRMPRCRHGYFHVVNNDY) of the PL amino acid sequences from tomato, Arabidopsis, strawberry and banana, comparatively analysed by Clustal X (Thompson et al., 2002). (c) Expression patterns of the tomato PL genes obtained from the TomExpress platform.
Detailed expression patterns of SlPL in wild‐type (WT) plants and hormone‐treated fruit
To assess the potential roles of SlPL throughout tomato development, we conducted detailed quantitative real‐time PCR (qRT‐PCR) to examine its transcription in different tissues. The expression levels were lower in roots (R), stems (S) and leaves (L) but relatively high in flowers (F). During tomato fruit development, expression of SlPL was relatively low in immature green (IMG) fruit and was negligible in mature green (MG) fruit. However, the expression level was drastically enhanced from the breaker stage (Br) and reached a maximum at 4 d after Br (Br+4) (Figure 2a). The observations were basically consistent with the data obtained from the TomExpress platform, further implying a possible crucial role of SlPL during fruit ripening/softening. Therefore, we isolated the total RNA from exocarp, sarcocarp, columella and locular tissue from fruit at Br+4 stage for qPCR analysis. The results showed that the SlPL expression level was relatively high in exocarp and sarcocarp (Figure 2b), indicating that SlPL functioned mainly in pericarp. To clarify the regulation of ripening‐related phytohormones on softening‐related genes, we tested the expression of SlPL in MG fruit of wild type (WT), respectively, treated with 1‐aminocyclopropane‐1‐carboxylic acid (ACC), ethephon (Eth), indole‐3‐acetic acid (IAA) and abscisic acid (ABA). The mRNA levels were significantly up‐regulated especially after the Eth treatment (Figure 2c). The results demonstrated that different types of phytohormones affected not only ripening but softening as well.
Figure 2.
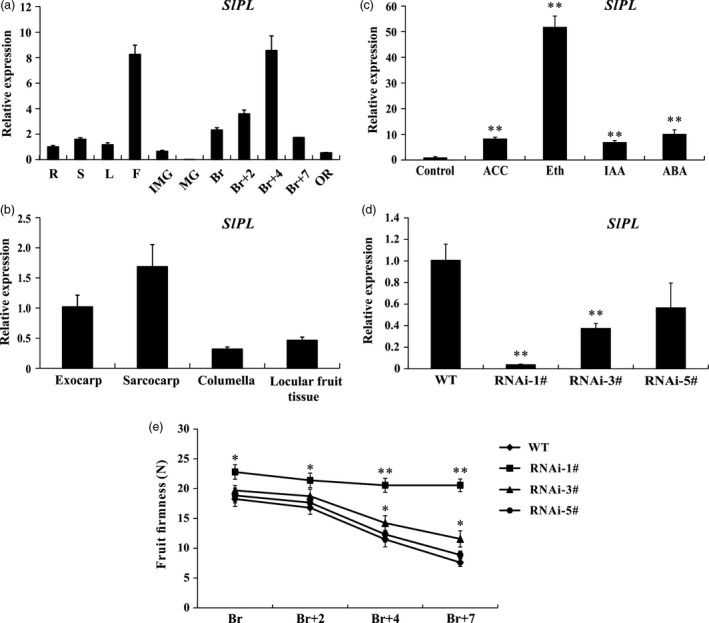
Expression patterns of SlPL (Solyc03g111690) and phenotype identification of SlPL‐RNAi transgenic tomato. (a) Expression of SlPL in various tissues including roots (R), stems (S), fully expanded leaves (L), flowers (F) and fruit at different developmental stages: immature green (IMG), mature green (MG), breaker (Br), 2 day after Br (Br+2), 4 day after Br (Br+4), 7 day after Br (Br+7, RR) and over ripe (OR). (b) Spatial expression of SlPL in WT fruit at Br+4 stage. (c) Response of SlPL to several ripening‐related hormones after treatment for 96 h. ACC, 1‐aminocyclopropane‐1‐carboxylic acid; Eth, ethephon; IAA, indole‐3‐acetic acid; and ABA, abscisic acid. (d) SlPL mRNA level in fruit of three independent RNAi lines (RNAi‐1#, RNAi‐3# and RNAi‐5#). (e) Phenotype of enhanced fruit firmness at different fruit development stages in RNAi lines. Quantitative PCR data represent mean values for three independent biological replicates (n = 3). The fruit firmness values represent the means ± standard error (SE) of 20 fruit per line at each stage. * and ** represent significant differences between SlPL‐RNAi lines and WT by t‐test with P < 0.05 and P < 0.01, respectively.
Down‐regulation of SlPL enhances fruit firmness
To further elucidate the function of SlPL, an RNAi construct targeting the fragment of this gene was created and transformed into WT tomato cv. Micro‐Tom plants via Agrobacterium tumefaciens mediated T‐DNA transfer. Real‐time PCR results showed that SlPL transcripts were significantly decreased in the transgenic lines compared with WT (Figure 2d). We next measured the fruit firmness at four different ripening stages: breaker (Br), 2 day after breaker (Br+2), Br+4 and 7 day after breaker (Br+7) (Figure 2e). The firmness in WT fruit decreased gradually with fruit ripening, with values of 18.3, 16.8, 11.5 and 7.6 newton (N) for each stage, respectively. In SlPL‐RNAi lines, it seems that the fruit firmness variation was deeply associated with the SlPL transcripts because no difference was observed for RNAi‐5# compared with WT, which had a transcriptional inhibition rate of 40%. For RNAi‐3#, with transcriptional inhibition rate of about 60%, the firmness was moderately enhanced. For RNAi‐1#, with nearly 95% inhibition of SlPL, the values of fruit firmness of 22.8, 21.4, 20.6 and 20.4 N for Br, Br+2, Br+4 and Br+7, respectively, showed minimum reductions in firmness throughout the experiment.
Silencing of SlPL leads to compact cells in fruit pericarp
As the pericarp is important in determining the rate of expansion and mechanical support for the whole fruit (Bargel and Neinhuis, 2005), histological analysis of the inner surface of the epidermis was conducted to compare the cell walls in WT and transgenic fruit. Increased cell separation was observed in SlPL‐RNAi fruit compared with WT (Figure 3a), and the corresponding statistical data were shown in Figure 3c. Also, the epidermal cells were smaller in SlPL‐RNAi transgenic fruit, leading to a larger number of epidermal cells (Figure 3a and c). Although cuticle is synthesized by the epidermal cell layer of the fruit pericarp, histological sectioning of sectors from fruit epicarp showed no difference in cuticle structure and thickness between WT and SlPL‐RNAi lines (Figure 3b and c). However, cell sizes and number as well as cell layer patterning of the whole pericarp changed. Consistent with the transverse sections, photographs revealed compact cells and increased cell numbers in exocarp, mesocarp and endocarp of RNAi fruit compared with WT (Figure 3d). Intriguingly, the phenomenon of ‘compact cells’ was reinforced in mesocarp, indicating that the site of action for SlPL was mostly in mesocarp cells rather than other cell types. These results suggest that the SlPL gene was involved in pericarp cell wall rearrangement during fruit softening.
Figure 3.
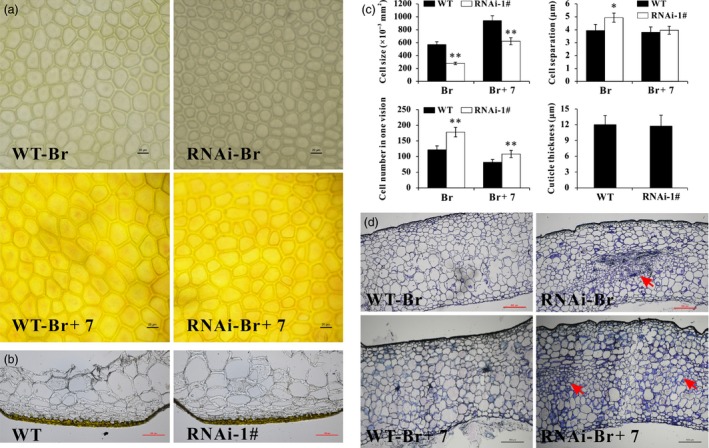
Differences of fruit pericarp cells in SlPL‐RNAi lines and WT. (a) Epidermal cells at the inner surface at Br and Br+7 stages from identical positions of different fruit. (b) Micrographs of unstained fruit epicarp sections from ripe tomatoes of WT and SlPL‐RNAi lines for the visualization of the cuticle. Bars = 100 μm. (c) Statistical analysis regarding epidermal cell size, cell number, cell separation and cuticle thickness of SlPL‐RNAi and WT fruit. Data are shown as means ± SE. * and ** indicate significant differences between SlPL‐RNAi lines and WT with P < 0.05 and P < 0.01, respectively, as determined by t‐test. (d) Paraffin transverse sections of pericarp tissues at Br and Br+7 stage fruit stained with toluidine blue. The differences in mesocarp tissues in SlPL‐RNAi fruit were indicated by red arrows. Sections were isolated from the same location in all fruit.
The SlPL‐RNAi fruit exhibits longer shelf‐life
The WT and SlPL‐RNAi fruit were harvested at Br+7 and stored at 23–25 °C with 55%–60% relative humidity until they reached complete deterioration. WT fruit were wrinkled after 6–7 day of storage, whereas SlPL‐RNAi fruit showed similar symptoms of senescence after 10–15 day of storage. At the time that effusion of juice and loss of texture and integrity occurred in WT fruit, the texture and fruit firmness were retained in SlPL‐RNAi fruit. Comparative analysis of images of fruit after storage of 0, 20 and 40 day showed less wrinkles for SlPL‐RNAi than WT fruit (Figure 4a). SlPL‐RNAi fruit exhibited lower physiological loss of water (PLW) than WT after 20 day of storage, although PLW did not differ in the first 20 day of storage (Figure 4b). During storage, fruit firmness was much higher in SlPL‐RNAi than in WT (Figure 4c).
Figure 4.
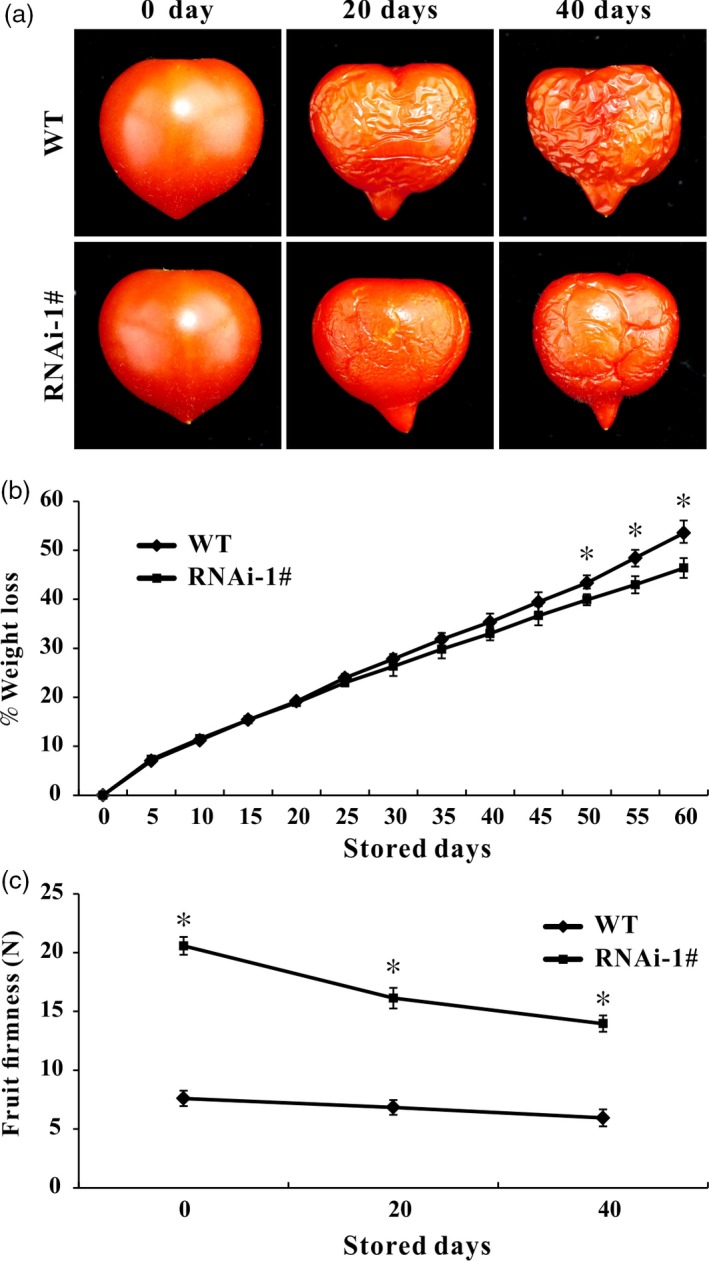
Suppression of SlPL extended fruit shelf‐life. (a) Red ripe (RR) fruit from SlPL‐RNAi lines and WT after storage at room temperature (23–25 °C and 55%–60% relative humidity) for 0, 20 and 40 day. (b) Physiological loss of water (PLW) in SlPL‐RNAi and WT fruit. The weight loss per fruit was calculated every 5 day during 0–60 day after storage. Values represent means ± SE (n = 20). * indicates significant differences between SlPL‐RNAi and WT fruit with P < 0.05, as determined by t‐test. (c) Change of fruit firmness in RNAi lines and WT. The fruit firmness per fruit was measured after storage of 0, 20 and 40 day. Average values were calculated for 20 individual fruit, and standard error of mean (SEM) was marked out. * indicates significant differences between SlPL‐RNAi and WT fruit with P < 0.05, as determined by t‐test.
SlPL‐RNAi fruit with reduced susceptibility to B. cinerea
Botrytis cinerea (B05.10) is the causal agent of grey mould disease, which is one of the most important postharvest pathogens of tomatoes (Williamson et al., 2007). When intact tomato fruit was sprayed with B. cinerea spore suspension, the WT fruit showed severe symptoms of infection with visible mycelium at 5 day postinoculation (dpi). However, the extent of infection was less severe in SlPL‐RNAi fruit (Figure 5a). When wounded fruit were inoculated with B. cinerea spore suspension, the size of the lesions did not increase at 1 dpi, indicating that establishment of the fungus needed about 24 h after inoculation. From 2 dpi, the spread of the infected area was greater in WT fruit. At 3 dpi, the average size of lesions in SlPL‐RNAi fruit was significantly less than that in WT, indicating reduced susceptibility to B. cinerea infection (Figure 5b). The lesion diameters at 1, 2 and 3 dpi in WT were nearly twice that in SlPL‐RNAi fruit (Figure 5c). The qPCR confirmed significantly more B. cinerea growing on WT fruits (Figure 5d). After invasion by B. cinerea, the expression of pathogen‐related genes (PR‐1b and PR‐1a) were down‐regulated in WT, while in RNAi fruit, their expressions were initially suppressed but significantly increased at 3 dpi. Notably, the mRNA levels were always higher in RNAi than WT fruit at all times after infection (Figure 5e and f).
Figure 5.
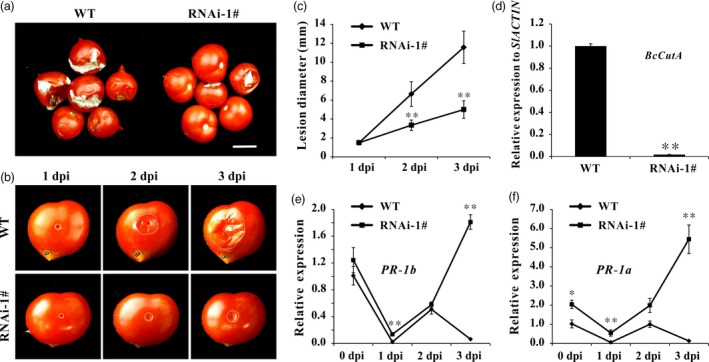
Suppression of SlPL in tomato decreased pathogen susceptibility. (a) Symptoms of intact WT and SlPL‐RNAi fruit after spraying of spores of B. cinerea (B05.10). Compared to rapid softening and collapsing symptoms in WT, fruit of the SlPL silenced line showed little mycelium at 5 days postinoculation (dpi). The scale bar represents 1 cm. (b) Symptoms of wounded WT and SlPL‐RNAi fruit after inoculation with B. cinerea (B05.10) at 1, 2 and 3 dpi. White dots represent the lesion margins. (c) Lesion diameter was measured at 1, 2 and 3 dpi. Error bars show the SE of means (n = 6). ** refers to significant differences between RNAi‐1# and WT with P < 0.01 determined by t‐test. (d) Quantitative PCR revealed more B. cinerea growing on the WT than on SlPL‐RNAi fruit at 3 dpi. The B. cinerea growth was calculated by comparing the ratio of B. cinerea DNA to tomato DNA. (e) Relative expression level of PR‐1b in RNAi‐1# and WT fruit (Br+14) after inoculation with B. cinerea at 0, 1, 2 and 3 dpi. (f) Relative expression level of PR‐1a in RNAi‐1# and WT fruit (Br+14) after inoculation with B. cinerea at 0, 1, 2 and 3 dpi. For qPCR analysis, the data represent mean values for three independent biological replicates. * and ** indicate significant differences between SlPL‐RNAi lines and WT with P < 0.05 and P < 0.01, respectively, as determined by t‐test.
Altered cell wall components and enhanced antioxidant capacity by silencing SlPL
The main components of plant cell walls are cellulose and pectin. In this study, contents of cellulose, hemicellulose, total pectin and water‐soluble pectin in WT and SlPL‐RNAi fruit were measured to determine whether the inhibition of SlPL led to changes in cell wall material. Three ripening stages were chosen for data collection: Br, Br+4 and Br+7. At each stage, the cellulose content in SlPL‐RNAi fruit (275.5, 218.5 and 200.3 mg/g, respectively) was higher than that in WT (231.1, 212.2 and 176.6 mg/g, respectively) (Figure 6a). The hemicellulose content in SlPL‐RNAi fruit (10.851 and 7.444 mg/g, respectively) was also greater than in WT (7.708 and 6.656 mg/g, respectively) at Br and Br+4 stages (Figure 6b). In contrast, the water‐soluble pectin was significantly reduced in SlPL‐RNAi fruit (Figure 6c), suggesting that most pectin in SlPL‐RNAi fruit cell walls was covalently attached to the wall matrix, and that down‐regulation of SlPL inhibited pectin degradation. Due to the decrease of water‐soluble pectin, the SlPL‐RNAi fruit also showed a slight reduction in total pectin (Figure 6d).
Figure 6.
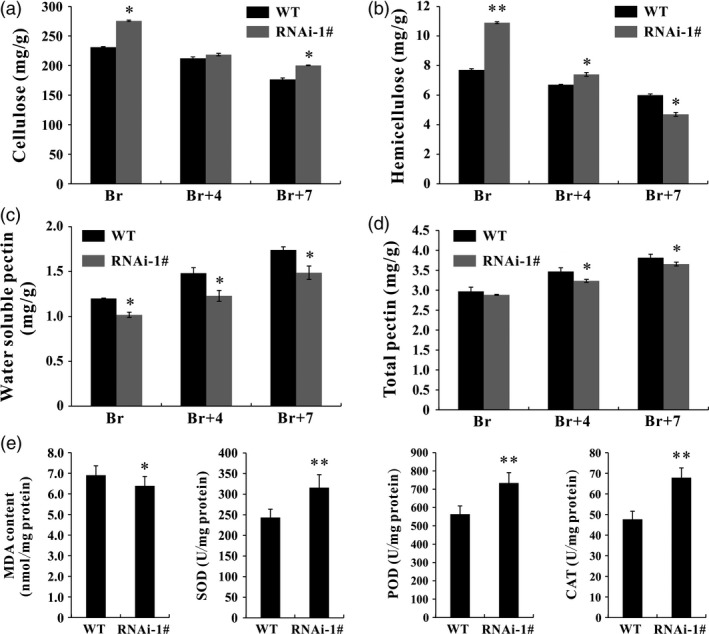
Pericarp cell wall components and antioxidant capacity in SlPL‐RNAi and WT fruit. (a) Content of cellulose (mg/g) in SlPL‐RNAi and WT fruit at Br, Br+4 and Br+7 stages. (b) Content of hemicellulose (mg/g) in SlPL‐RNAi and WT fruit at Br, Br+4 and Br+7 stages. (c) Content of water‐soluble pectin (WSP) (mg/g) in SlPL‐RNAi and WT fruit at Br, Br+4 and Br+7 stages. (d) Content of total pectin (mg/g) in SlPL‐RNAi and WT fruit at Br, Br+4 and Br+7 stages. (e) The malondialdehyde (MDA) level (nmol/mg protein) and antioxidant enzyme activity of superoxide dismutase (SOD), peroxidase (POD) and catalase (CAT) (U/mg protein) in SlPL‐RNAi and WT fruit at Br+14 stage. The error bars represent the SE calculated from three measurements for each cell wall material. * and ** indicate significant differences between SlPL‐RNAi and WT plants with P < 0.05 and P < 0.01, respectively, as determined by t‐test.
Malondialdehyde (MDA) is a by‐product of lipid peroxidation and can be used to evaluate the damage by oxidative stress during tissue senescence. Superoxide dismutase (SOD), peroxidase (POD) and catalase (CAT) are three important antioxidant enzymes. MDA level, together with SOD, POD and CAT activities, was monitored in fresh red ripe (RR) fruit. Compared with WT, there were lower MDA levels and higher SOD, POD and CAT enzyme activities in SlPL‐RNAi fruit (Figure 6e), suggesting that it had stronger antioxidant ability.
Differentially expressed genes (DEGs) based on RNA sequencing data
Gene expression profiles of WT and SlPL‐RNAi fruit were compared using RNA sequencing (RNA‐Seq) data at Br+4 stage. A twofold difference in expression level was set as the threshold for data significance. In RNAi fruit, 943 genes consisting of 181 up‐regulated and 762 down‐regulated genes showed apparent differential expression. The DEGs were functionally identified using gene ontology (GO) analysis and categorized into 11 main categories (Figure 7a and b). In addition, using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, many DEGs were found to be involved in cell wall metabolism and plant–pathogen interaction (Figure 7c and d). Exhaustive analysis showed that 49 DEGs were phytohormone related, including 22 with ethylene, 14 with auxin, four with gibberellins, five with abscisic acid, two with jasmonate acid, one with salicylic acid and one with cytokinins (Table S2). Transcription factors (TFs) are an important part of functional genomics and participate in various regulatory networks in higher plants. We searched the RNA‐Seq data and found 68 DEGs encoding TFs. The dominant TF families were the ERF (16 DEGs), bHLH (7 DEGs), MADS‐box (7 DEGs), WRKY (7 DEGs), AP2 (5 DEGs), bZIP (4 DEGs), B3 (4 DEGs), MYB (3 DEGs), NAC (2 DEGs), C2H2 (2 DEGs) and GRAS (1 DEG) (Table S3), suggesting important roles of these TFs in fruit ripening/softening. Moreover, functional annotation revealed that many DEGs were involved in cell wall modification (63 DEGs), oxidative stress (39 DEGs) and pathogen resistance (78 DEGs) (Tables S4 and S5). The transcriptomic data partly explained the molecular basis underlying the extended shelf‐life and pathogen resistance of the SlPL‐RNAi fruit.
Figure 7.
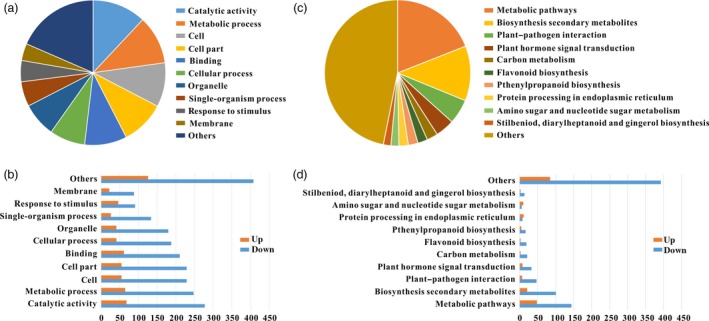
Classification of DEGs with RNA‐Seq data. (a) GO classification and functional annotation of the 943 genes showing more than twofold differences in expression between RNA‐1# and WT fruit at Br+4 stage. (b) Functional classification of all differentially expressed genes at Br+4 stage with GO classification. (c) Pathway and KEGG classification and functional annotation of the 943 genes showing more than twofold differences in expression between RNA‐1# and WT fruit at Br+4 stage. (d) Functional classification of all differentially expressed genes at Br+4 stage with pathway and KEGG classification.
To validate our transcriptomic data, 29 genes were chosen for supplementary qRT‐PCR analysis in WT and RNAi fruits (Figure 8). The selected genes comprised five phytohormone related, eight TF, eight cell wall‐processing and eight stress‐related genes. The qRT‐PCR results were highly consistent with transcriptomic data for all tested genes.
Figure 8.
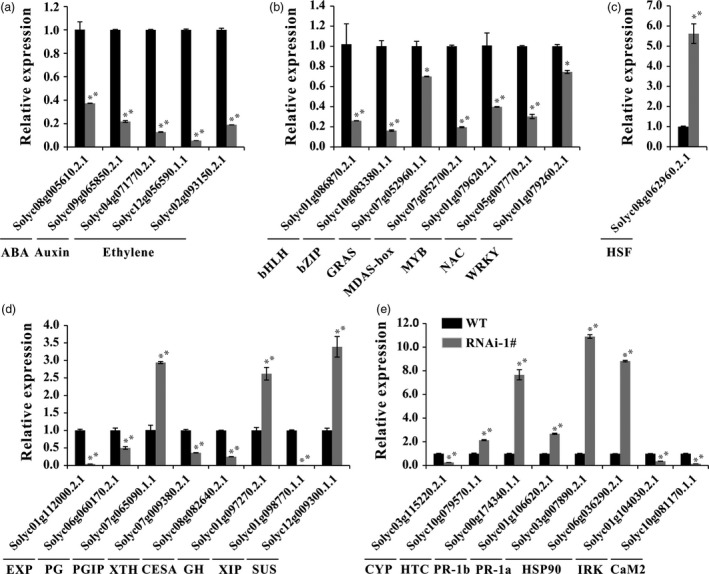
Verification of selected DEGs by real‐time PCR. (a) Relative expression of genes related to phytohormones ABA, auxin and ethylene. (b) and (c) Relative expression of genes related to transcription factors. (d) Relative expression of genes related to cell wall modification and degradation. (e) Relative expression of genes related to oxidative stress and pathogen resistance. Error bars show the SE between three biological replicates performed (n = 3). * and ** indicate P < 0.05 and P < 0.01, respectively.
Discussion
Fruit softening is a result of degradation/modification of cell wall polymers by the action of cell wall associated hydrolases (Brummell and Harpster, 2001; Fisher and Bennet, 1991). However, many enzymes cannot be exploited to genetically control the softening of fruit. In the past few decades, pectate lyase has been documented as a potential candidate for manipulating fruit softening in several fruit crops, including strawberry, banana, mango and grape (Chourasia et al., 2006; Marín‐Rodríguez et al., 2002; Payasi and Sanwal, 2003). Very recently, Uluisik et al. (2016) revealed that suppressing a PL gene in tomato could enhance fruit firmness and extend shelf‐life without impacting the flavour, aroma, taste and nutritional value. In the present study, we identified 22 members of the PL gene family in tomato and divided them into five subfamilies (Figure 1a). Using a RNAi strategy, we further characterized a ripening/softening‐specific gene SlPL (Solyc03 g111690). Due to the high homology of sequences (over 80%) between transgenic construction and several other PL genes expressed in immature fruit (SlPL16, SlPL5 and SlPL19), qPCR analysis was performed at different development stages: IMG, MG and Br+4. The results demonstrated that these genes were affected to some extent at one or two stages, while SlPL was severely inhibited at all stages especially during fruit ripening (Figure S1). The expression of other two genes SlPL12 (Solyc05 g014000) and SlPL15 (Solyc06 g071840) which were closed to SlPL in phylogenetic tree also showed no significant suppression at RR fruit (Figure S1e). These findings indicated that SlPL was most relevant to the above phenotype while other gene family members might have made minor contributions. In agreement with the work by Uluisik et al. (2016), the SlPL‐RNAi lines were capable of producing fruit with enhanced firmness and extended shelf‐life (Figures 2 and 4) without negative effects on vegetative growth, fruit development, days to maturity, and fruit yield except for a decreased weight of seeds and ratio of seed germination (Table S6).
Fruit softening is usually accompanied by a reduction in cell turgor, due to increasing concentrations of solutes in the cell wall space and wall loosening (Shackel et al., 1991). A tomato mutant cultivar ‘delayed fruit deterioration’ (DFD) showed minimal transpirational water loss and substantially elevated cellular turgor likely due to specific compositional or ultrastructural characteristics of the fruit cuticle (Saladié et al., 2007). Although the cuticle can function as an important barrier to reduce water loss and extend shelf‐life (Segado et al., 2016), it showed very similar structure and thickness between WT and RNAi fruit in our research (Figure 3b and c). This result is in accordance with the report by Uluisik et al. (2016) in which the SlPL‐RNAi fruit showed longer shelf‐life. In their study, the cuticle waxes of SlPL‐RNAi and WT fruit did not significantly differ, although genes involved in epidermal regulation and cuticle development such as PROTODERMAL FACTOR 2‐like (Solyc06g035940) and CER1 (Solyc03g065250) were up‐regulated (Uluisik et al., 2016). Additionally, Giménez et al. (2015) demonstrated that greater thickness of the cuticle may not affect the breaking stress. Generally, an increase in cell size is indicative of a positive turgor pressure that facilitates cell expansion. However, anatomical observations showed that the size of epidermal cells was reduced in RNAi fruit (Figure 3), suggesting that the enhanced fruit firmness in SlPL‐RNAi transgenic lines was not caused by cell expansion. The main effect of SlPL is in the tricellular junction zones of pericarp parenchyma cells, and it breaks down the cross‐linked homogalacturonan (HG) polymers in both middle lamella and tricellular junctions (Uluisik et al., 2016). Consistently, we found more compact parenchyma cells in RNAi fruit pericarps than in WT (Figure 3d). In addition, silencing SlPL would inhibit the eliminative cleavage of de‐esterified pectin. Compared with WT, contents of cellulose and hemicelluloses were significantly higher in RNAi fruit, while the water‐soluble pectin and total pectin were lower (Figure 6).
Botrytis cinerea is a necrotrophic fungal pathogen capable of killing plant cells and proliferating on dead tissues (Prins et al., 2000). Simultaneous suppression of LePG and LeExp1 in ripening tomato fruit reduced wall disassembly, slowed fruit softening and decreased ripening‐associated susceptibility to B. cinerea (Cantu et al., 2008). Unfortunately, suppression of LePG or LeExp1 alone did not reduce the susceptibility of fruit to B. cinerea infection, suggesting that PG and Exp may act cooperatively against B. cinerea invasion in tomato (Brummell et al., 1999, 2002; Kalamaki et al., 2003; Powell et al., 2003). It is remarkable that the SlPL‐RNAi fruit in our study showed reduced susceptibility to B. cinerea (Figure 5). These findings support the ability of SlPL in specific control of fruit softening and B. cinerea invasion. As the expression of SlPL5 was also up‐regulated during fruit ripening in WT and co‐suppressed in PL‐RNAi fruit (Figure S1), we monitored expression of SlPL and SlPL5 after B. cinerea infection, the trend of SlPL expression in RNAi fruit after inoculation was similar to that in WT. However, the mRNA level of SlPL5 showed no significant change at 2 or 3 dpi in RNAi fruit after B. cinerea treatment (Figure S2), demonstrating that SlPL may have contributed to the observed pathogen resistance rather than SlPL5. Actually, pectate lyase can activate defence systems by releasing plant cell wall oligogalacturonides, which can then function as defence elicitors (De Lorenzo et al., 1991). Del/Ros1 tomato fruit showed decreased B. cinerea susceptibility due to higher anthocyanin accumulation, which contributed to attenuated oxidative damage (Zhang et al., 2013). SOD, POD and CAT are important antioxidant enzymes serving as efficient scavengers of reactive oxygen species to avoid oxidative damage. The SlPL‐RNAi RR fruit possessed lower MDA content and higher antioxidant enzyme activities compared with WT fruit (Figure 6e).
In our RNA‐Seq data, 943 genes showed more than twofold difference in expression level at Br+4 stage compared with WT. This result differed from the recent study, which showed fewer than 120 genes were altered in transcriptomes at the orange stage (Uluisik et al., 2016). This difference may be because Uluisik et al. used a different cultivar. Additionally, our results showed suppression of SlPL5 and SlPL19 (Figure S1). The latter was highly expressed in immature fruit of WT. These gene expression changes could be also responsible for the differences between our findings and those reported by Uluisik et al. (2016) with respect to cell size and differential gene expression. The DEGs were associated with plant hormones, fruit ripening, cell wall degradation, as well as pathogens. Ethylene, IAA and ABA are important hormones related to fruit ripening, to which the softening‐related gene SlPL can respond (Figure 2c). Moreover, the hormone‐related DEGs were also found in SlPL‐RNAi fruit (Figure 8). Genes encoding polygalacturonase (SlPG) and xyloglucan endotransglycosylase (SlXTH), and other genes involved in cell wall softening, showed substantially lower expression in SlPL‐RNAi fruit during ripening (Figure 8). Although the silencing of individual genes might only have minor effects on softening, the simultaneous suppression of a number of cell wall modification enzymes would likely result in significant reductions in the rate of fruit softening (Giovannoni, 2004; Goulao and Oliveira, 2008; Powell et al., 2003).
In conclusion, we demonstrated that SlPL was undeniably an important pectinase related to cell wall disassembly and fruit softening in tomato. The extended shelf‐life and disease resistance of SlPL‐RNAi fruit revealed the great value of pectate lyase.
Materials and methods
Plant materials and growth conditions
Solanum lycopersicum cv. Micro‐Tom was selected as the wild type (WT). Tomato plants were grown in a greenhouse with standard conditions (16/8 h and 25 °C/18 °C of day/night, 80% humidity and 250 μmol/m2/s light intensity) and watered daily. The tissues of roots (R); stems (S); leaves (L); flowers (F); IMG fruit; MG fruit; Br stage fruit; yellow fruit, Br+2; orange fruit, Br+4; RR fruit, Br+7; Br+14; and over ripe fruit (OR) were collected from WT and transgenic tomato plants. The tissues of exocarp, sarcocarp, columella and locular fruit tissues of WT Br+4 stage fruit were also collected. For RNA extraction and cell wall composition analysis, samples were collected, frozen in liquid nitrogen immediately and stored at –80 °C. For cytological, texture and postharvest biology analyses, experiments were performed using fresh fruit material.
Tomato PL gene family analysis and expression data mining
In the SOL Genomics Network (http://solgenomics.net), ‘pectate lyase’ was inputted in the annotation text to search unigene families. Multiple sequences alignments of the amino acid sequences of PL proteins from different species were analysed by ClustalX 1.83. A phylogenetic tree was constructed using MEGA6 software, and statistical confidence of the nodes of the tree was calculated using 10 000 bootstrap replicates. Expression patterns of 22 tomato PL genes during vegetative and reproductive development were predicted with TomExpress bioinformatics platform (http://gbf.toulouse.inra.fr/tomexpress).
Vector construction and plant transformation
For RNAi vector construction, 377‐basepairs DNA sequences of SlPL were selected, and the sense and antisense orientation fragments were then amplified with primers listed in Table S1. Then, the amplified products were digested with EcoR1/Kpn1 and XbaI/BamH1, respectively, and linked into the pHANNIBAL vector (Wesley et al., 2001). Finally, the double‐stranded RNA expression unit, cauliflower mosaic virus 35S promoter and OCS terminator were inserted into the plant binary vector pBIN19 with SacI and XbaI restriction sites. The SlPL‐RNAi binary plasmid was transferred into Agrobacterium strain GV3101, and Agrobacterium‐mediated transformation was performed following the protocols described by Fillatti et al. (1987). Transgenic plants were identified by PCR with primers of neomycin phosphotransferase II (NPTII) (Table S1). The positive transgenic plants were selected and used for subsequent experiments.
RNA isolation and qPCR analysis
Total RNAs were extracted using Trizol reagent following the protocol provided by the manufacturer (Invitrogen) and treated with DNase I (Thermo Scientific). About 2 μg of total RNA from each sample was used for first‐strand cDNA synthesis. For qPCR, the reaction was performed using SyBR Green PCR Master Mix (CWBIO, China). The procedures of PCR amplification consisted of an initial incubation at 95 °C for 20 s, followed by 40 cycles of 95 °C for 3 s and 60 °C for 30 s. PCR products were monitored using Bio‐Rad CFX connect (Bio‐Rad). Each sample was amplified in triplicate, and normalization was performed by comparative CT method. SlActin (Solyc03g078400) was selected as the internal reference. All primers for qPCR are listed in Table S1.
Treatments of tomato WT MG fruit with different hormones
For hormone treatment experiments, tomato fruit at MG stage was used. Final concentration of 100 μm ACC, IAA, ABA and 0.4% ethephon in solution (pH 5.6) containing 10 mm MES and 3% sorbitol (all Sigma‐Aldrich products) was prepared in the above order, and buffer injection was performed according to a preliminary study (Orzaez et al., 2006). After treatment, fruit were incubated in a culture room at 26 °C, under 16/8‐h light/dark cycle with a light intensity of 100 μmol/m2/s. Pericarp was collected after 96 h and frozen at –80 °C for further analysis.
Fruit texture analysis
Fruit firmness was determined based on compression mass and skin puncture strength of fresh intact fruit collected at different fruit ripening stage, including MG, Br, Br+2, Br+4 and Br+7 stages, using a GY‐4 digital fruit sclerometer (Aiwoshi, China). The analyser was equipped with a circular probe of diameter 7 mm, speed 10 mm/s and depression 4 mm, recorded in newton (N). Twenty fruit from different plants were taken at each stage, and each fruit was tested twice at equidistant points along the equatorial plane of the fruit. Values represent means ± standard error (SE; n = 20). For postharvest fruit firmness analysis, 20 fruit were used at each storage stage for measurement.
Cytological assessment of fruit pericarp
Fruit epidermis was carefully isolated from the fruit surface with colourless nail enamel and tweezers. The detached segment was rinsed twice with distilled water before fixation on tissue‐bound slide and then mounted in 15% hydrochloric acid (HCl) under a cover slip. Observation was performed with the inner surface of epidermis on a Nikon microscope (Nikon Co. Ltd., Japan). Six exocarp slices from identical positions of different fruit were isolated at Br and Br+7 (RR) stages. For each exocarp slice, epidermal cell size (not including cell wall), cell separation and cell number (in one field of vision below a 40× object lens) were measured at three different positions using ImageProPlus software.
For cytological assessment of cuticle and fruit cells from exocarp to endocarp, three fruit of different plants were collected at the Br and Br+7 (RR) stages. Fresh pericarp sections were prepared and fixed in FAA solution (5% formaldehyde, 5% acetic acid and 50% ethanol) to examine pericarp cell wall structure. Fixed tomato fruit pericarp was dehydrated in ethanol series up to 95%, cleared with histo‐clear xylene and embedded in paraffin wax at 58 °C. Paraffin‐embedded transverse sections of 8 μm thickness were cut by microtome and attached to glass slides, and sections were dewaxed with xylene and stained with 0.05% toluidine blue. Micrographs of unstained fruit epicarp sections from RR tomato fruits were chosen for cuticle observation. A Nikon microscope was used, and cuticle thickness was estimated using ImageProPlus software
Shelf‐life analysis
For determination of shelf‐life, fruit at the RR (Br+7) stage were detached and surface sterilized first, and then, each fruit was placed in a plastic jar kept at room temperature (23–25 °C and 55%–60% relative humidity). Every 5 day, fresh weight of each fruit was measured to calculate the PLW, and visual softening and collapse of fruit were assessed (Nambeesan et al., 2010). Twenty fruit were taken from different plants for analysis.
Botrytis cinerea infection
The B. cinerea (B05.10) was grown on 1% potato dextrose agar, and conidia were harvested from sporulating colonies and filtered through glass wool as described by Stefanato et al. (2009). Surface sterilized fruit (Br+14) collected from WT and SlPL‐RNAi lines were sprayed thoroughly with spores (2.5 × 105 spores/mL) five times in a fume cabinet and kept at 25 °C, in high humidity conditions. Infection symptoms were observed at 5 dpi. For wound inoculation, the fungal culture was diluted with sterile water to 5 × 104 spores/mL and inoculated at 25 °C for 1.5 h to stimulate germination. Next, 5 μL of spore suspension was added to each wound. Lesion diameter was measured at 24, 48 and 72 h postinoculation. To quantify B. cinerea growth, 1 cm3 of fruit tissue around the lesion area was collected at 72 h postinoculation and total DNA extracted using a Plant Genomic DNA Kit (CWBIO, China). Purified DNA (50 ng) was used for qPCR as described by Zhang et al. (2013).
Cell wall content analysis
For measurement of cellulose content, 100 mg of pericarp was ground with liquid nitrogen and digested with 60–70 mL cold 60% (V/V) sulphuric acid (H2SO4) for 1.5 h on ice. Next, filtration was performed and 2 mL of the filtered solution was mixed with 0.5 mL of 2% anthrone in a test tube, and concentrated H2SO4 was gently poured in along the tube wall. Subsequently, the reaction mixture was water‐bathed at 100 °C for 10 min and taken out for measurement at a wavelength of 620 nm, with pure cellulose used as a standard.
For analysis of hemicellulose content, hydrolysis of hemicellulose was conducted with a water‐bath at 100 °C for 45 min with 2 mol/L HCl. After centrifuging, the supernatant was transferred to a 100‐mL volumetric flask, which was then neutralized to rose pink using sodium hydroxide solution with phenolphthalein as the pH indicator. The solution was subsequently diluted and filtrated. Finally, the main content of hemicellulose, pentosan was measured using lichen phenol–HCl.
Cell wall polysaccharides were first prepared as alcohol‐insoluble solids (AIS) (Rosli et al., 2004). Afterwards, 50 mg of AIS was suspended in 50 mL of distilled water and incubated at 50 °C for 30 min under continuous stirring, and then, the solution was filtered and the residue washed thrice with 5 mL of distilled water. The pooled filtrate was denoted as WSP. The residue was extracted with 0.5 m H2SO4 at 100 °C for 1 h and denoted as HSP. Both WSP and HSP were analysed according to the m‐hydroxydiphenyl method (Blumenkrantz and Ashobe‐Hansen, 1973) using D‐galacturonic acid (D‐GA) as a standard. For each sample, tissue was pooled from six individual fruit. Three independent experiments were carried out and results expressed as mean ± SD of all replicates.
Lipid peroxidation and antioxidant enzyme assays
Fresh fruit (Br+14) of SlPL‐RNAi and WT were homogenized and extracted with 0.1 mol/L (pH7.8) cold phosphate buffer saline on ice. Afterwards, the SOD activity was assessed according to Beauchamp and Fridovich (1971). POD activity was assessed by measuring the increase in absorbance at 470 nm due to the formation of tetraguaiacol (Sakharov, 1993). The CAT activity was measured according to Aebi (1974). MDA content was estimated according to Heath and Packer (1968).
RNA‐Seq analysis for DEGs
RNA‐Seq was carried out by BGI Life Tech Co. Ltd. (Wuhan, China). Total RNA was isolated from Br+4 stage fruit using RNeasy Plant Mini Kit (Tiangen, China) following the manufacturer's protocol. Samples from both RNAi and WT fruit were performed with two biological replications. The cDNA libraries were constructed using a TruSeq™ RNA Sample Preparation Kit (Illumina) and subsequently sequenced using an Illumina HiSeq™ 2000 platform. The quality of RNA‐Seq data was assessed with the RSeQC‐2.3.2 program (http://code.google.com/p/rseqc/). Clean reads obtained from the raw reads were obtained by removing the adapter and low quality sequences and then mapped to the annotated genome sequence of S. lycopersicum in the Tomato Sol Genomic Network database (http://solgenomics.net/). Transcript abundance was also normalized by the fragments per kilobase of exon per million mapped reads (FRKM) method, and Cuffdiff software (http://cufflinks.cbcb.umd.edu/) was used to identify DEGs. A twofold difference in expression levels was set as the threshold for the determination of the significant changes. GO functional enrichment and KEGG pathway analysis were carried out with goatools (https://github.com/tanghaibao/goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home. do). All RNA‐Seq data were available in Data S2 online.
Statistical analysis
All experiments were repeated three times independently, and all results are reproducible. Statistical results are presented as means ± standard error. To compare group differences, two‐tailed Student's t‐tests were used. P values less than 0.05 were recognized as significant.
Supporting information
Figure S1 Relative expression of SlPL and other highly homologous genes in WT and SlPL‐RNAi fruit.
Figure S2 Relative expression of SlPL and SlPL5 in WT and SlPL‐RNAi fruit with the inoculation of B. cinerea (B05.10).
Table S1 Details of gene primers used in this article.
Table S2 The DEGs involved in phytophormone metabolism and signaling transduction.
Table S3 The DEGs encoding the members of TF families.
Table S4 Expression analysis of DEGs involved in cell wall modification.
Table S5 Expression analysis of DEGs involved in oxidative stress and pathogen resistance.
Table S6 Evaluation of WT and RNAi plants for agronomic characteristics.
Data S1 The accession numbers and according amino acid sequences of 22 tomato PLs, 26 Arabidopsis PLs and two softening‐related PLs from strawberry and banana.
Data S2 The RNA‐Seq original data.
Acknowledgements
This work was supported by the National Key Research and Development Program (2016YFD0400101), the National Basic Research Program of China (2013CB127106), the National Natural Science Foundation of China (31572175), and the Committee of Science and Technology of Chongqing (cstc2014kjcxljrc0020). The authors declare no conflict of interest.
References
- Aebi, H. (1974) Catalase In Methods of Enzymatic Analysis (Bergmeyer H.U., ed), pp. 673–677. New York: Academic Press. [Google Scholar]
- Bargel, H. and Neinhuis, C. (2005) Tomato (Lycopersicon esculentum Mill.) fruit growth and ripening as related to the biomechanical properties of fruit skin and isolated cuticle. J. Exp. Bot. 413, 1049–1060. [DOI] [PubMed] [Google Scholar]
- Beauchamp, C. and Fridovich, I. (1971) Superoxide dis‐mutase: improved assays and an assay applicable to acrylamide gels. Anal. Biochem. 44, 276–287. [DOI] [PubMed] [Google Scholar]
- Blumenkrantz, N. and Ashobe‐Hansen, G. (1973) New method for quantitative determination of uronic acids. Anal. Biochem. 54, 484–489. [DOI] [PubMed] [Google Scholar]
- Brummell, D.A. and Harpster, M.H. (2001) Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol. Biol. 47, 311–340. [PubMed] [Google Scholar]
- Brummell, D.A. , Harpster, M.H. , Civello, P.M. , Palys, J.M. , Bennett, A.B. and Dunsmuir, P. (1999) Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell, 11, 2203–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell, D.A. , Howie, W.J. , Ma, C. and Dunsmuir, P. (2002) Postharvest fruit quality of transgenic tomatoes suppressed in expression of a ripening‐related expansin. Postharvest Biol. Tec. 25, 209–220. [Google Scholar]
- Cantu, D. , Vicente, A.R. , Greve, L.C. , Dewey, F.M. , Bennett, A.B. and Labavitch, J.M. (2008) The intersection between cell wall disassembly, ripening, and fruit susceptibility to botrytis cinerea . Proc. Natl Acad. Sci. USA, 105, 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita, N.C. and Gibeaut, D.M. (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3, 1–30. [DOI] [PubMed] [Google Scholar]
- Cassab, G.I. and Varner, J.E. (1988) Cell wall proteins. Ann. Rev. Plant Physiol. Mol. Biol. 39, 321–353. [DOI] [PubMed] [Google Scholar]
- Chourasia, A. , Sane, V.A. and Nath, P. (2006) Differential expression of pectate lyase during ethylene‐induced postharvest softening of mango (Mangifera indica var. Dashehari). Physiol. Plantarum. 128, 546–555. [Google Scholar]
- De Lorenzo, G. , Cervone, F. , Hahn, M.G. , Darvill, A. and Albersheim, P. (1991) Bacterial endopectate lyase: evidence that plant cell wall pH prevents tissue maceration and increases the half‐life of elicitor‐active oligogalacturonides. Physiol. Mol. Plant P. 39, 335–344. [Google Scholar]
- De Silva, J. , Arrowsmith, D.A. , Whiteman, S. and Robinson, S. (1994) Xyloglucan endotransglycosylase and plant growth. J. Exp. Bot. 45, 1693–1701. [Google Scholar]
- Domínguez ‐Puigjaner, E. , LLop, I. , Vendrell, M. and Prat, S.A. (1997) A cDNA clone highly expressed in ripe banana fruit shows homology to pectate lyases. Plant Physiol. 114, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillatti, J.A.J. , Kiser, J. , Rose, R. and Comai, L. (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary agrobacterium tumefaciens vector. Nat. Biotechnol. 5, 726–730. [Google Scholar]
- Fisher, R.L. and Bennet, A.B. (1991) Role of cell wall hydrolases in fruit ripening. Annu. Rev. Plant Biol. 42, 675–703. [Google Scholar]
- Giménez, E. , Dominguez, E. , Pineda, B. , Heredia, A. , Moreno, V. , Lozano, R. and Angosto, T. (2015) Transcriptional activity of the mads box arlequin/tomato agamous‐like1 gene is required for cuticle development of tomato fruit. Plant Physiol. 168, 1036–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni, J.J. (2001) Molecular biology of fruit maturation and ripening. Annu. Rev. Plant Physiol. Mol. Biol. 52, 725–749. [DOI] [PubMed] [Google Scholar]
- Giovannoni, J.J. (2004) Genetic regulation of fruit development and ripening. Plant Cell, 16, S170–S180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulao, L.F. and Oliveira, C.M. (2008) Cell wall modifications during fruit ripening: when a fruit is not the fruit. Trends Food Sci. Tech. 19, 4–25. [Google Scholar]
- Grierson, D. and Schuch, W. (1993) Control of ripening. Philoso. T. R. Soc. B. 342, 241–250. [Google Scholar]
- Hall, L.N. , Tucker, G.A. , Smith, C.J.S. , Watson, C.F. , Seymour, G.B. , Bundick, Y. , Boniwell, J.M. et al (1993) Antisense inhibition of pectin esterase gene expression in transgenic tomatoes. Plant J. 3, 121–129. [Google Scholar]
- Harholt, J. , Suttangkakul, A. and Vibe Scheller, H. (2010) Biosynthesis of pectin. Plant Physiol. 153, 384–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, R.L. and Packer, L. (1968) Photoperoxidation in isolated chloroplasts.I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 125, 189–198. [DOI] [PubMed] [Google Scholar]
- Huber, D.J. (1984) Strawberry fruit softening: the potential roles of polyuronides and hemicelluloses. J. Food Sci. 47, 1310–1315. [Google Scholar]
- Jiménez‐Bermúdez, S. , Redondo‐Nevado, J. , Munoz‐Blanco, J. , Caballero, J.L. , Lopez‐Aranda, J.M. , Valpuesta, V. , Pliego‐Alfaro, F. et al (2002) Manipulation of strawberry fruit softening by antisense expression of a pectate lyase gene. Plant Physiol. 128, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamaki, M.S. , Powell, A.L. , Struijs, K. , Labavitch, J.M. , Reid, D.S. and Bennett, A.B. (2003) Transgenic overexpression of expansin influences particle size distribution and improves viscosity of tomato juice and paste. J. Agr. Food Chem. 51, 7465–7471. [DOI] [PubMed] [Google Scholar]
- Knee, M. , Sargent, J.A. and Osborne, D.J. (1977) Cell wall metabolism in developing strawberry fruits. J. Exp. Bot. 28, 377–396. [Google Scholar]
- Langley, K.R. , Martin, A. , Stenning, R. , Murray, A.J. , Hobson, G.E. , Schuch, W.W. and Bird, C.R. (1994) Mechanical and optical assessment of the ripening of tomato fruit with reduced polygalacturonase activity. J. Sci. Food Agr. 66, 547–554. [Google Scholar]
- Lunn, D. , Phan, T.D. , Tucker, G.A. and Lycett, G.W. (2013) Cell wall composition of tomato fruit changes during development and inhibition of vesicle trafficking is associated with reduced pectin levels and reduced softening. Plant Physiol. Bioch. 66, 91–97. [DOI] [PubMed] [Google Scholar]
- Marín‐Rodríguez, M.C. (2001) Investigation of the role of pectate lyase in banana fruit softening. PhD thesis, University of Greenwich.
- Marín‐Rodríguez, M.C. , Orchard, J. and Seymour, G.B. (2002) Pectate Lyases, cell wall degradation and fruit softening. J. Exp. Bot. 53, 2115–2119. [DOI] [PubMed] [Google Scholar]
- Medina‐Escobar, N. , CaÂrdenas, J. , Moyano, E. , Caballero, J.L. and MunÄoz‐Blanco, J. (1997) Cloning molecular characterisation and expression pattern of a strawberry ripening‐specific cDNA with sequence homology to pectate lyase from higher plants. Plant Mol. Biol. 34, 867–877. [DOI] [PubMed] [Google Scholar]
- Medina‐Suárez, R. , Manning, K. , Fletcher, J. , Aked, J. , Bird, C.R. and Seymour, G.B. (1997) Gene expression in the pulp of ripening bananas. Plant Physiol. 115, 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meli, V.S. , Ghosh, S. , Prabha, T.N. , Chakraborty, N. , Chakraborty, S. and Datta, A. (2010) Enhancement of fruit shelf life by N‐glycan processing enzymes. Proc. Natl Acad. Sci. USA, 6, 2413–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambeesan, S. , Datsenka, T. , Ferruzzi, M.G. , Malladi, A. , Mattoo, A.K. and Handa, A.K. (2010) Overexpression of yeast spermidine synthase impacts ripening, senescence and decay symptoms in tomato. Plant J. 63, 836–847. [DOI] [PubMed] [Google Scholar]
- Nunan, K.J. , Davies, C. , Robinson, S.P. and Fincher, G.B. (2001) Expression patterns of cell wall‐modifying enzymes during grape berry development. Planta, 214, 257–264. [DOI] [PubMed] [Google Scholar]
- Orzaez, D. , Mirabel, S. , Wieland, W.H. and Granell, A. (2006) Agroinjection of tomato fruits. A tool for rapid functional analysis of transgenes directly in fruit. Plant Physiol. 140, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payasi, A. and Sanwal, G.G. (2003) Pectate lyase activity during ripening of banana fruit. Phytochemistry, 63, 243–248. [DOI] [PubMed] [Google Scholar]
- Pilatzke‐Wunderlich, I. and Nessler, C.L. (2001) Expression and activity of cell‐wall‐degrading enzymes in the latex of opium poppy, Papaver somniferum L. Plant Mol. Biol. 45, 567–576. [DOI] [PubMed] [Google Scholar]
- Powell, A.L.T. , Kalamaki, M.S. , Kurien, P.A. , Gurrieri, S. and Bennett, A.B. (2003) Simultaneous transgenic suppression of LePG and LeExp1 influences fruit texture and juice viscosity in a fresh market tomato variety. J. Agr. Food Chem. 51, 7450–7455. [DOI] [PubMed] [Google Scholar]
- Prins, T.W. , Tudzynski, P. , Tiedemann, A.V. , Tudzynski, B. , Have, A.T. , Hansen, M.E. , Tenberge, K. et al (2000) Infection Strategies of Botrytis cinerea and related necrotrophic pathogens In Fungal Pathology, vol. 2 (Kronstad J.W., ed), pp. 33–64. Dordrecht Netherland: Springer, Netherlands. [Google Scholar]
- Redgwell, R.J. , MacRae, E. , Hallett, J. , Ficher, M. , Perry, J. and Harker, R. (1997) In vivo and in vitro swelling of cell‐walls during fruit ripening. Planta, 203, 162–173. [Google Scholar]
- Rose, J.K.C. , Lee, H.H. and Bennett, A.B. (1997) Expression of a divergent expansin gene is fruits specific and ripening‐regulated. Pro. Natl. Acad. Sci. USA, 94, 5955–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosli, H.G. , Civello, P.M. and Martinez, G.A. (2004) Changes in cell wall composition of three Fragariax ananassa cultivars with different softening rate during ripening. Plant Physiol. Bioch. 42, 823–831. [DOI] [PubMed] [Google Scholar]
- Sakharov, J.G. (1993) Oxygen stress and superoxide dismutases. Plant Physiol. 101, 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saladié, M. , Matas, A.J. , Isaacson, T. , Jenks, M.A. , Goodwin, S.M. , Niklas, K.J. , Xiaolin, R. et al (2007) A reevaluation of the key factors that influence tomato fruit softening and integrity. Plant Physiol. 144, 1012–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago‐Doménech, N. , Jiménez‐Bemúdez, S. , Matas, A.J. , Rose, J.K.C. , Munoz‐Blanco, J. , Mercado, J.A. and Quesada, M.A. (2008) Antisense inhibition of a pectate lyase gene supports a role for pectin depolymerization in strawberry fruit softening. J. Exp. Bot. 59, 2769–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segado, P. , Domínguez, E. and Heredia, A. (2016) Ultrastructure of the epidermal cell wall and cuticle of tomato fruit (Solanum lycopersicum L.) during development. Plant Physiol. 170, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackel, K.A. , Greve, C. , Labavitch, J.M. and Ahmadi, H. (1991) Cell turgor changes associated with ripening in tomato pericarp tissue. Plant Physiol. 97, 814–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy, R.E. , Kramer, M. and Hiatt, W.R. (1988) Reduction of polygalacturonase activity in tomato fruit by antisense RNA. Pro. Natl Acad. Sci. USA, 85, 8805–8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A.P. , Pandey, S.P. , Rajluxmi Pandey, S. , Nath, P. and Sane, A.P. (2011) Transcriptional activation of a pectate lyase gene, RbPel1, during petal abscission in rose. Postharvest Biol. Tec. 60, 143–148. [Google Scholar]
- Smith, C.J. , Watson, C.F. , Morris, P.C. , Bird, C.R. , Seymour, G.B. , Gray, J.E. , Arnold, C. et al (1990) Inheritance and effect on ripening of antisense polygalacturonase genes in transgenic tomatoes. Plant Mol. Biol. 14, 369–379. [DOI] [PubMed] [Google Scholar]
- Smith, D.L. , Abbott, J.A. and Gross, K.C. (2002) Down‐regulation of tomato beta‐galactosidase 4 results in decreased fruit softening. Plant Physiol. 129, 1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanato, F.L. , Abou‐Mansour, E. , Buchala, A. , Kretschmer, M. , Mosbach, A. , Hahn, M. , Bochet, C.G. et al (2009) The ABC transporter BcatrB from Botrytis cinerea exports camalexin and is a virulence factor on Arabidopsis thaliana . Plant J. 58, 499–510. [DOI] [PubMed] [Google Scholar]
- Taniguchi, Y. , Ono, A. , Sawatani, M. , Nanba, M. , Kohno, K. , Usui, M. , Kurimoto, M. et al (1995) Cry j I, a major allergen of Japanese cedar pollen, has a pectate lyase enzyme activity. Allergy, 50, 90–93. [DOI] [PubMed] [Google Scholar]
- Thompson, J. , Gibson, T. and Higgins, D. (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics, 2, 1–22. [DOI] [PubMed] [Google Scholar]
- Tieman, D.M. and Handa, A.K. (1994) Reduction in pectin methylesterase activity modifies tissue integrity and cation levels in ripening tomato (Lycopersicon esculentum Mill.) fruits. Plant Physiol. 106, 429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman, D.M. , Harriman, R.W. , Ramamohan, G. and Handa, A. K. (1992) An antisense pectin methylesterase gene alters pectin chemistry and soluble solids in tomato fruit. Plant Cell, 4, 667–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uluisik, S. , Chapman, N.H. , Smith, R. , Poole, M. , Adams, G. , Gillis, R.B. , Besong, T.M.D. et al (2016) Genetic improvement of tomato by targeted control of fruit softening. Nat. Biotechnol. (online) 34(9), 950–952 https://doi.org/10.1038/nbt.3602. [DOI] [PubMed] [Google Scholar]
- Wen, B. , Ström, A. , Tasker, A. , West, G. and Tucker, G.A. (2013) Effect of silencing the two major tomato fruit pectin methylesterase isoforms on cell wall pectin metabolism. Plant Biology, 15, 1025–1032. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V. , Helliwell, C.A. , Smith, N.A. , Wang, M. , Rouse, D.T. , Liu, Q. , Gooding, P.S. et al (2001) Construct design for efficient, effective and high‐throughput gene silencing in plants. Plant J. 27, 581–590. [DOI] [PubMed] [Google Scholar]
- Willats, W.G.T. , McCartney, L. , Mackey, W. and Knox, J.P. (2001) Pectin: cell biology and prospects for functional analysis. Plant Mol. Biol. 47, 9–27. [PubMed] [Google Scholar]
- Williamson, B. , Tudzynski, B. , Tudzynski, P. and Van Kan, J.A.L. (2007) Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8, 561–580. [DOI] [PubMed] [Google Scholar]
- Wing, R.A. , Yamaguchi, J. , Larabell, S.K. , Ursin, V.M. and McCormick, S. (1989) Molecular and genetic characterization of two pollen‐expressed genes that have sequence similarity to pectate lyases of the plant pathogen Erwinia . Plant Mol. Biol. 14, 17–28. [DOI] [PubMed] [Google Scholar]
- Wu, Y. , Qiu, X. , Du, S. and Erickson, L. (1996) PO149, a new member of pollen pectate lyase‐like gene family from alfalfa. Plant Mol. Biol. 32, 1037–1042. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Butelli, E. , De Stefano, R. , Schoonbeek, H.J. , Magusin, A. , Pagliarani, C. , Wellner, N. et al (2013) Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr. Biol. 23, 1094–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Relative expression of SlPL and other highly homologous genes in WT and SlPL‐RNAi fruit.
Figure S2 Relative expression of SlPL and SlPL5 in WT and SlPL‐RNAi fruit with the inoculation of B. cinerea (B05.10).
Table S1 Details of gene primers used in this article.
Table S2 The DEGs involved in phytophormone metabolism and signaling transduction.
Table S3 The DEGs encoding the members of TF families.
Table S4 Expression analysis of DEGs involved in cell wall modification.
Table S5 Expression analysis of DEGs involved in oxidative stress and pathogen resistance.
Table S6 Evaluation of WT and RNAi plants for agronomic characteristics.
Data S1 The accession numbers and according amino acid sequences of 22 tomato PLs, 26 Arabidopsis PLs and two softening‐related PLs from strawberry and banana.
Data S2 The RNA‐Seq original data.


