Figure 2. USP14 is a DUB for PER.
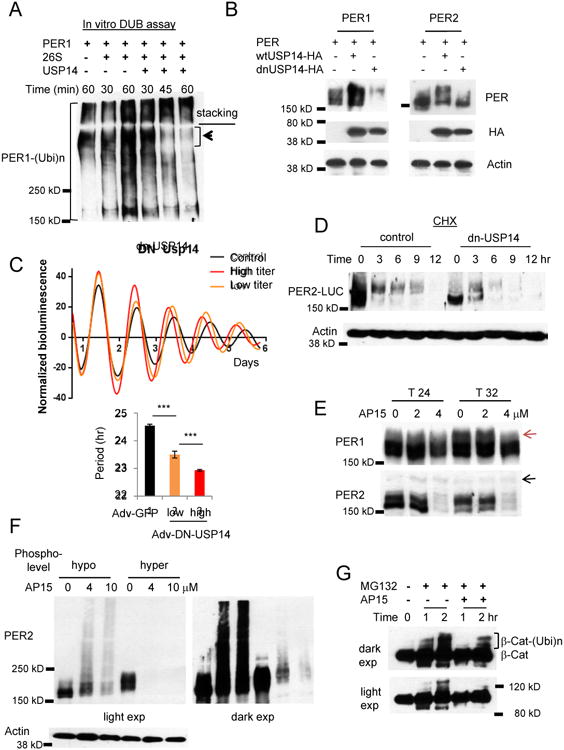
(A) In vitro deubiquitination assays are consistent with the model that polyubiquitinated PER1 is deubiquitinated and degraded by a 26S proteasome-USP14 complex. Note that PER1 species at the top of the resolving gel disappeared when co-incubated with the complex. (B) PER is destabilized when USP14 activity is inhibited. Per1 or Per2 was co-transfected with wt Usp14 or a dnUsp14 in HEK293a cells. (C) The circadian clock is accelerated when dnUSP14 is expressed. Per2Luc MEFs were infected with two different titers of dnUsp14-adenovirus. Data in the lower graph are mean+/-SEM (n=8 each). Representative of three experiments. (D) Hypophosphorylated PER2 species are predominant and rapidly degraded in dnUSP14-overexpressing cells. Representative of two experiments. (E) Nuclear PER species are predominantly affected by the deubiquitinase inhibitor drug b-AP15. Note that hyperphosphorylated PER1 species (indicated by the red arrow) accumulate at T32 only in control and 2 μM-treated cells, but not in 4 μM-treated cells. The black arrow indicated a nonspecific band. T24 and T32 hours represent times after a 2-hr serum shock to synchronize cell rhythms. (F) The sensitivity of hyperphosphorylated PER species is reproduced with overexpressed PER. Hypo- and hyperphosphorylated PER2 was inducibly overexpressed in Rosa-TA; tetO-Per2 MEFs. (G) Ubiquitination of β-Cat is delayed in b-AP15 treated cells. See also Figures S2D-G.
