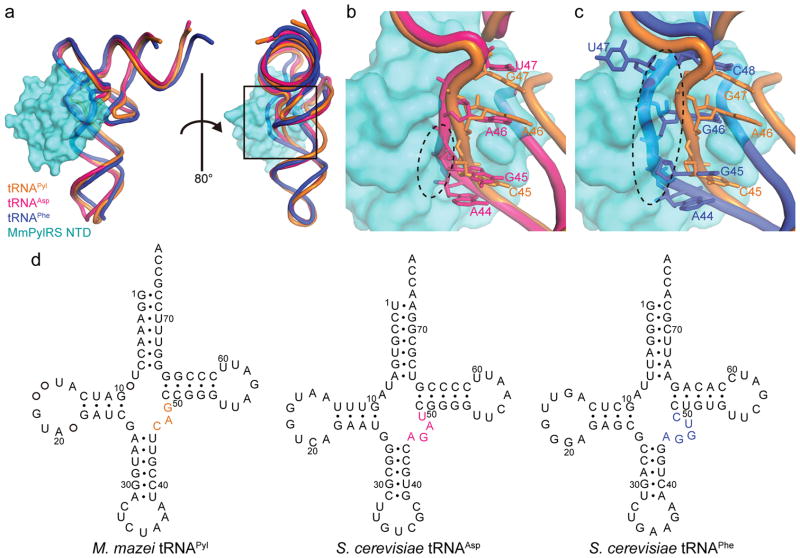
Figure 2. Structural basis for the PylRS specificity to tRNAPyl.
(a) Superposition of the tRNAAsp (PDB ID: 2TRA) and tRNAPhe (PDB ID: 1EHZ) structures onto the tRNAPyl of the MmPylRS NTD•tRNAPyl complex was achieved by overlaying the phosphorus atoms of the acceptor stem, anticodon stem, and T–stem/loop (40 atoms for tRNAAsp, rmsd=3.04, and 41 for tRNAPhe, rmsd=3.21). MmPylRS NTD is shown as a surface model. (b) Close-up view of the variable loops of tRNAAsp and tRNAPyl (A44-U47and C45-G47, respectively). (c) Close-up view of the variable loops of tRNAPhe and tRNAPyl (A44-C48 and C45-G47, respectively). The steric clash between the MmPylRS NTD and the canonical tRNA is represented by a black dashed circle. Posttranscriptional modifications are omitted for clarity. (d) The cloverleaf structures of M. mazei tRNAPyl, S. cerevisiae tRNAAsp, and S. cerevisiae tRNAPhe show the different sizes of the variable loops.