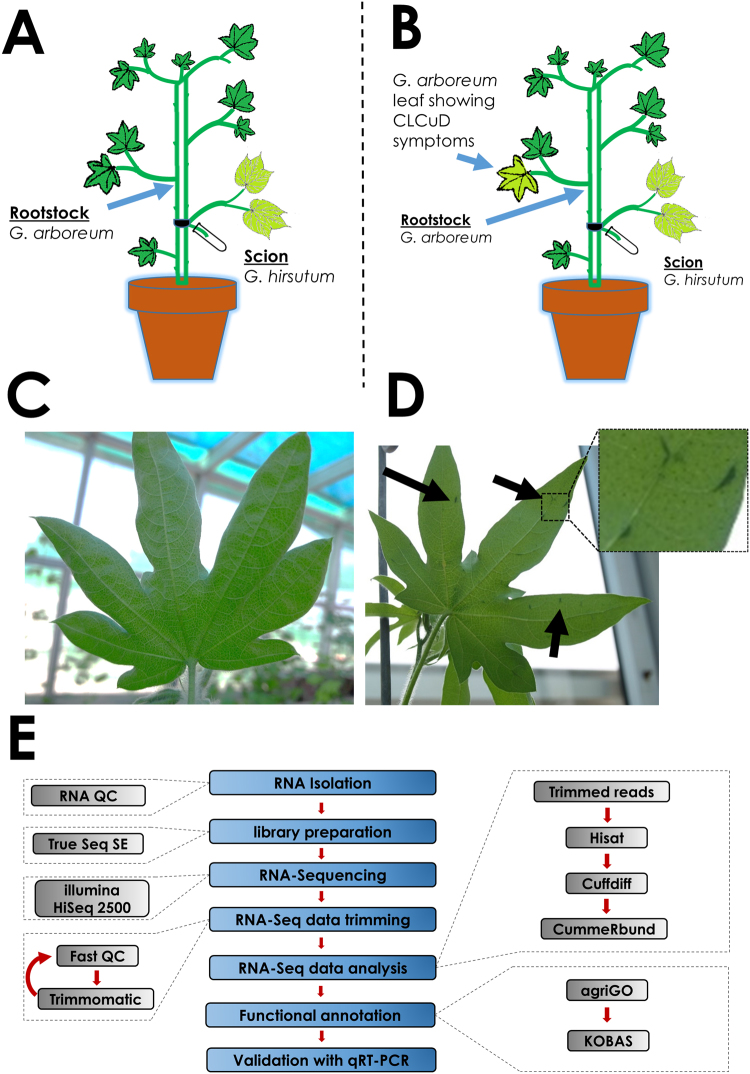
Figure 1.
Experimental design, methodology and RNA-Seq pipeline used in study. Panel A and B show the graphical representation of grafting experiments (A) scion of cotton leaf curl disease (CLCuD) infected Gossypium hirsutum was used on a rootstock of G arboreum; asymptomatic leaves of CLCuD were collected for RNA extraction and sequencing. (B) Leaves showing symptoms of CLCuD were collected for RNA extraction and sequencing. (C) Shows an asymptomatic CLCuD free leaf of G. arboreum. (D) Shows a leaf with very mild symptoms of CLCuD including vein swelling and darkening, highlighted with black arrows. (E) Shows the workflow of the RNA-Seq experiment and the tools used at each step.