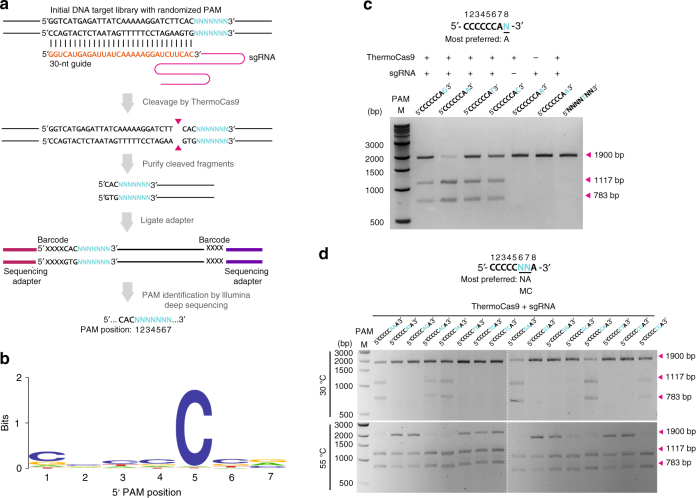
Fig. 2.
ThermoCas9 PAM analysis. a Schematic illustrating the in vitro cleavage assay for discovering the position and identity (5′-NNNNNNN-3′) of the protospacer adjacent motif (PAM). Magenta triangles indicate the cleavage position. b Sequence logo of the consensus 7 nt long PAM of ThermoCas9, obtained by comparative analysis of the ThermoCas9-based cleavage of target libraries. Letter height at each position is measured by information content. c Extension of the PAM identity to the eighth position by in vitro cleavage assay. Four linearized plasmid targets, each containing a distinct 5′-CCCCCCAN-3′ PAM, were incubated with ThermoCas9 and sgRNA at 55 °C for 1 h, then analyzed by agarose gel electrophoresis. Supplementary Fig. 8 shows the uncropped gel image. d In vitro cleavage assays for DNA targets with different PAMs at 30 and 55 °C. Sixteen linearized plasmid targets, each containing one distinct 5′-CCCCCNNA-3′ PAM, were incubated with ThermoCas9 and sgRNA, then analyzed for cleavage efficiency by agarose gel electrophoresis. See also Supplementary Fig. 3. Supplementary Fig. 9 shows the uncropped gel images