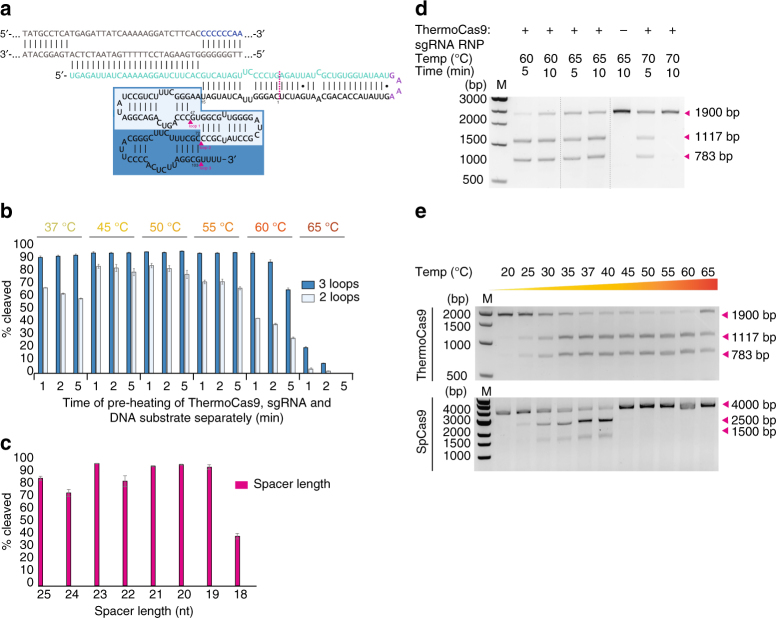
Fig. 3.
ThermoCas9 temperature range and effect of sgRNA-binding. a Schematic representation of the sgRNA and a matching target DNA. The target DNA, the PAM and the crRNA are shown in gray, blue and turquoise, respectively. The site where the crRNA is linked with the tracrRNA is shown in purple. The dark blue and light blue boxes indicate the predicted three and two loops of the tracrRNA, respectively. The 41-nt truncation of the repeat-anti-repeat region and the three loops of the sgRNA are indicated by the magenta dotted line and magenta triangles, respectively. b The importance of the predicted three stem-loops of the tracrRNA scaffold was tested by transcribing truncated variants of the sgRNA and evaluating their ability to guide ThermoCas9 to cleave target DNA at various temperatures. Average values of three replicates are shown, with error bars representing S.D. The blots of one of the replicates are shown in Supplementary Fig. 10. c The importance of the length of the spacer was tested by transcribing truncated variants of the initial spacer in the sgRNA and evaluating their ability to guide ThermoCas9 to cleave target DNA at 55 °C. Average values of three replicates are shown, with error bars representing S.D.. The blots of one of the replicates are shown in Supplementary Fig. 11. d To identify the maximum temperature, endonuclease activity of ThermoCas9:sgRNA RNP complex was assayed after incubation at 60, 65, and 70 °C for 5 or 10 min. The pre-heated DNA substrate was added and the reaction was incubated for 1 h at the corresponding temperature. e Comparison of active temperature range of ThermoCas9 and SpCas9 by activity assays conducted after 5 min of incubation at the indicated temperature. The pre-heated DNA substrate was added and the reaction was incubated for 1 h at the same temperature